-Search query
-Search result
Showing 1 - 50 of 434 items for (author: graham & s)

EMDB-29907: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v); consensus map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29908: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), locally refined map
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29909: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gat: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gau: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gav: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-42940: 
Dimer of Hendra virus prefusion F trimers
Method: single particle / : Byrne PO, Blade EG, McLellan JS

EMDB-42481: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42482: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D

EMDB-42483: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-2heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42485: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5
Method: single particle / : Park YJ, Veesler D

EMDB-42486: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D

PDB-8ur5: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-1heptad-I53_dn5 (local refinement of TH-1heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8ur7: 
I53_dn5 nanoparticle displaying the trimeric HA heads with heptad domain, TH-6heptad-I53_dn5 (local refinement of TH-6heptad)
Method: single particle / : Park YJ, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-16657: 
Cryo-EM structure of the fd bacteriophage capsid major coat protein pVIII
Method: helical / : Boehning J, Bharat TAM

PDB-8ch5: 
Cryo-EM structure of the fd bacteriophage capsid major coat protein pVIII
Method: helical / : Boehning J, Bharat TAM

EMDB-28743: 
Cryo-EM structure of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2
Method: single particle / : Duan HD, Li H

EMDB-28748: 
Cryo-EM structure of the S. cerevisiae guanine nucleotide exchange factor Gea2
Method: single particle / : Duan HD, Li H

PDB-8ezj: 
Cryo-EM structure of the S. cerevisiae Arf-like protein Arl1 bound to the Arf guanine nucleotide exchange factor Gea2
Method: single particle / : Duan HD, Li H

PDB-8ezq: 
Cryo-EM structure of the S. cerevisiae guanine nucleotide exchange factor Gea2
Method: single particle / : Duan HD, Li H

EMDB-28728: 
Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-28729: 
Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-28730: 
Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-41089: 
Cryo-EM structure of RSV preF in complex with Fab 2.4K
Method: single particle / : McCool RS, McLellan JS

PDB-8t7a: 
Cryo-EM structure of RSV preF in complex with Fab 2.4K
Method: single particle / : McCool RS, McLellan JS

EMDB-29280: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B

PDB-8flj: 
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA
Method: single particle / : Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin C, Buyukyoruk M, Neselu K, Eng ET, Lander GC, Wiedenheft B
EMDB-41299: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Method: single particle / : Nygaard R, Mancia F
EMDB-41303: 
Transmembrane map
Method: single particle / : Nygaard R, Mancia F
EMDB-41304: 
Periplasmic map
Method: single particle / : Nygaard R, Mancia F

EMDB-27566: 
Prefusion-stabilized Nipah virus fusion protein
Method: single particle / : Byrne PO, Blade EG, McLellan JS

EMDB-27577: 
Prefusion-stabilized Hendra virus fusion protein
Method: single particle / : Byrne PO, Blade EG, McLellan JS

EMDB-27590: 
Prefusion-stabilized Nipah virus fusion protein, dimer of trimers
Method: single particle / : Byrne PO, Blade EG, McLellan JS

PDB-8dng: 
Prefusion-stabilized Nipah virus fusion protein
Method: single particle / : Byrne PO, Blade EG, McLellan JS

PDB-8dnr: 
Prefusion-stabilized Hendra virus fusion protein
Method: single particle / : Byrne PO, Blade EG, McLellan JS

PDB-8do4: 
Prefusion-stabilized Nipah virus fusion protein, dimer of trimers
Method: single particle / : Byrne PO, Blade EG, McLellan JS
EMDB-29248: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
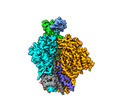
EMDB-29264: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
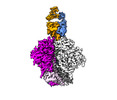
EMDB-29288: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fk5: 
Cryo-EM Structure of PG9RSH DU011 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8fl1: 
Cryo-EM Structure of PG9RSH DU025 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-8flw: 
Cryo-EM Structure of PGT145 DU303 Fab in complex with BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD
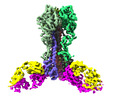
EMDB-27139: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8d21: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-16540: 
Neurofascin isoform NF155 extracellular domain
Method: single particle / : McKie SJ, Deane JE, Butt BG

EMDB-26652: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 4H3
Method: single particle / : Byrne PO, McLellan JS

EMDB-26658: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2D3
Method: single particle / : Byrne PO, McLellan JS

EMDB-26659: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8
Method: single particle / : Byrne PO, McLellan JS

EMDB-26660: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H1
Method: single particle / : Byrne PO, McLellan JS

EMDB-26662: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2B12
Method: single particle / : Byrne PO, McLellan JS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
