-Search query
-Search result
Showing all 34 items for (author: gonzalez & rl)

EMDB-25026: 
Anaphase Promoting Complex delta APC7
Method: single particle / : Ferguson CJ, Brown NG, Prabu JR, Watson ER, Schulman BA, Bonni A

EMDB-25027: 
Anaphase Promoting Complex delta APC7 + UBE2C,substrate,UbV,CDH1
Method: single particle / : Ferguson CJ, Brown NG, Prabu JR, Watson ER, Schulman BA, Bonni A

EMDB-23211: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

EMDB-23215: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

PDB-7l7f: 
Cryo-EM structure of human ACE2 receptor bound to protein encoded by vaccine candidate BNT162b1
Method: single particle / : Lees JA, Han S

PDB-7l7k: 
Cryo-EM structure of protein encoded by vaccine candidate BNT162b2
Method: single particle / : Lees JA, Han S

EMDB-11788: 
Cryo-EM structure of the RUVBL1-RUVBL2-DHX34 complex
Method: single particle / : Lopez-Perrote A, Rodriguez CF, Llorca O

EMDB-11789: 
RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34
Method: single particle / : Lopez-Perrote A, Rodriguez CF, Llorca O

EMDB-20772: 
Fab397 in complex with NPNA8 peptide (Class 1)
Method: single particle / : Ward AB, Torres JL

EMDB-20773: 
Fab397 in complex with NPNA8 peptide (Class 2)
Method: single particle / : Ward AB, Torres JL

EMDB-20774: 
Fab397 in complex with NPNA8 peptide (Class 3)
Method: single particle / : Ward AB, Torres JL
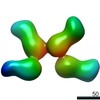
EMDB-20775: 
Fab397 in complex with rsCSP (Class 1)
Method: single particle / : Ward AB, Torres JL

EMDB-20776: 
Fab397 in complex with rsCSP (Class 2)
Method: single particle / : Ward AB, Torres JL
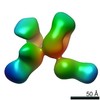
EMDB-20777: 
Fab397 in complex with rsCSP (Class 3)
Method: single particle / : Ward AB, Torres JL
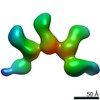
EMDB-20778: 
Fab397 in complex with rsCSP (Class 4)
Method: single particle / : Ward AB, Torres JL
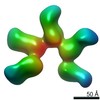
EMDB-20779: 
Fab397 in complex with rsCSP (Class 5)
Method: single particle / : Ward AB, Torres JL

EMDB-20780: 
Fab397 in complex with rsCSP (Class 6)
Method: single particle / : Ward AB, Torres JL

EMDB-0643: 
30S initiation complex
Method: single particle / : Frank J, Gonzalez Jr RL, kaledhonkar S, Fu Z, Caban K, Li W, Chen B, Sun M

EMDB-0661: 
70S Elongation Competent Ribosome
Method: single particle / : Frank J, Gonzalez Jr RL

EMDB-0662: 
70S initiation complex
Method: single particle / : Frank J, Gonzalez Jr RL, kaledhonkar S, Fu Z, Caban K, Li W, Chen B, Sun M

EMDB-3619: 
RnQV1-W1118 empty capsid
Method: single particle / : Mata CP, Luque D, Gomez-Blanco J, Rodriguez JM, Suzuki N, Ghabrial SA, Carrascosa JL, Trus BL, Caston JR

EMDB-3437: 
Three-dimensional reconstruction of W1075 strain Rosellinia necatrix quadrivirus 1
Method: single particle / : Luque D, Mata CP, Gonzalez-Camacho F, Gonzalez JM, Gomez-Blanco J, Alfonso C, Rivas G, Havens WM, Kanematsu S, Suzuki N, Ghabrial SA, Trus BL, Caston JR

EMDB-3438: 
Three-dimensional reconstruction of W1118 strain Rosellinia necatrix quadrivirus 1
Method: single particle / : Luque D, Mata CP, Gonzalez-Camacho F, Gonzalez JM, Gomez-Blanco J, Alfonso C, Rivas G, Havens WM, Kanematsu S, Suzuki N, Ghabrial SA, Trus BL, Caston JR

EMDB-3725: 
Negative-stain EM structure of the A. tumefaciens outer-membrane core complex
Method: single particle / : Costa TRD, Orlova EV, Waksman G

EMDB-4086: 
RNA polymerase I-Rrn3 complex
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

EMDB-4087: 
Monomeric RNA polymerase I at 5.6 A resolution
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

EMDB-4088: 
Monomeric RNA polymerase I at 4.9 A resolution
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

EMDB-5784: 
EttA-bound E. coli 70S ribosome complex containing P-site tRNA and A-site tRNA
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-5785: 
EttA-bound E. coli 70S ribosome complex containing P-site tRNA
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-5786: 
EttA-bound E. coli 70S ribosome complex
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-5841: 
EttA-bound E. coli 70S ribosome complex containing P-site tRNA and A-site tRNA (raw map)
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-5842: 
EttA-bound E. coli 70S ribosome complex containing P-site tRNA (raw map)
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-5843: 
EttA-bound E. coli 70S ribosome complex (raw map)
Method: single particle / : Chen B, Boel G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL, Hunt JF, Frank J

EMDB-2062: 
Cryphonectria nitschkei Virus (CnV)capsid
Method: single particle / : Gomez-Blanco J, Luque D, Gonzalez JM, Carrascosa JL, Alfonso C, Trus B, Havens WH, Ghabrial SA, Caston JR
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
