-Search query
-Search result
Showing 1 - 50 of 320 items for (author: gonen & m)

EMDB-43714: 
Cryo-EM structure of VP3-VP6 heterohexamer
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43716: 
Cryo-EM structure of BTV star-subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43719: 
Cryo-EM structure of BTV pre-subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43722: 
Cryo-EM structure of pre-subcore from in vitro assembled particles
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43723: 
Cryo-EM structure of BTV empty virion
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43724: 
Cryo-EM structure of BTV empty core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43725: 
Cryo-EM structure of BTV empty pre-core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43726: 
Cryo-EM structure of BTV subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43727: 
Cryo-EM structure of BTV virion
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43728: 
Subtomogram averaging of BTV virion in host cells
Method: subtomogram averaging / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43730: 
Cryo-EM structure of BTV core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

EMDB-43731: 
Cryo-EM structure of BTV pre-core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w12: 
Cryo-EM structure of VP3-VP6 heterohexamer
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w19: 
Cryo-EM structure of BTV star-subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w1c: 
Cryo-EM structure of BTV pre-subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w1i: 
Cryo-EM structure of BTV subcore
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w1o: 
Cryo-EM structure of BTV virion
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w1r: 
Cryo-EM structure of BTV core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8w1s: 
Cryo-EM structure of BTV pre-core
Method: single particle / : Xia X, Sung PY, Martynowycz MW, Gonen T, Roy P, Zhou ZH

PDB-8skw: 
MicroED structure of d(CGCGCG)2 Z-DNA
Method: electron crystallography / : Haymaker A, Bardin AA, Martynowycz MW, Nannenga BL

EMDB-28028: 
I3-01 in an expanded conformation
Method: single particle / : McCarthy S, Gonen S

EMDB-28029: 
I3-01 map refined in C1
Method: single particle / : McCarthy S, Gonen S

EMDB-40351: 
The MicroED structure of proteinase K crystallized by suspended drop crystallization
Method: electron crystallography / : Gillman C, Nicolas WJ, Martynowycz MW, Gonen T

PDB-8sdk: 
The MicroED structure of proteinase K crystallized by suspended drop crystallization
Method: electron crystallography / : Gillman C, Nicolas WJ, Martynowycz MW, Gonen T

EMDB-29587: 
MicroED structure of Proteinase K from lamellae milled from multiple plasma sources
Method: electron crystallography / : Martynowycz MW, Shiriaeva A, Clabbers MTB, Nicolas WJ, Weaver SJ, Hattne J, Gonen T

PDB-8fyo: 
MicroED structure of Proteinase K from lamellae milled from multiple plasma sources
Method: electron crystallography / : Martynowycz MW, Shiriaeva A, Clabbers MTB, Nicolas WJ, Weaver SJ, Hattne J, Gonen T

EMDB-27148: 
Zebrafish MFSD2A isoform B in inward open ligand bound conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27149: 
Zebrafish MFSD2A isoform B in inward open ligand-free conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27150: 
Zebrafish MFSD2A isoform B in inward open ligand 1A conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27151: 
Zebrafish MFSD2A isoform B in inward open ligand 1B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27152: 
Zebrafish MFSD2A isoform B in inward open ligand 2B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-27153: 
Zebrafish MFSD2A isoform B in inward open ligand 3C conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2s: 
Zebrafish MFSD2A isoform B in inward open ligand bound conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2t: 
Zebrafish MFSD2A isoform B in inward open ligand-free conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2u: 
Zebrafish MFSD2A isoform B in inward open ligand 1A conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2v: 
Zebrafish MFSD2A isoform B in inward open ligand 1B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2w: 
Zebrafish MFSD2A isoform B in inward open ligand 2B conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

PDB-8d2x: 
Zebrafish MFSD2A isoform B in inward open ligand 3C conformation
Method: single particle / : Nguyen C, Lei HT, Lai LTF, Gallentino MJ, Mu X, Matthies D, Gonen T

EMDB-15244: 
Tomogram of an Ebola VLP composed of GP, VP40, NP, VP24 and VP35 at pH 7.4 (Figure 1A-D)
Method: electron tomography / : Winter SL, Chlanda P
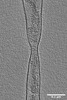
EMDB-15268: 
Tomogram of an Ebola VLP composed of VP40 at pH 4.5 (Figure 1J)
Method: electron tomography / : Winter SL, Chlanda P

EMDB-15951: 
Tomogram of an EBOV-infected Huh7 cell showing a late endosome with internalized EBOV particles
Method: electron tomography / : Winter SL, Chlanda P

EMDB-15956: 
Tomogram of an extracellular EBOV particle adjacent to an EBOV-infected Huh7 cell
Method: electron tomography / : Winter SL, Chlanda P

EMDB-16128: 
Tomogram of a late endosome of A549 cell infected with influenza A virus.
Method: electron tomography / : Klein S, Chlanda P

EMDB-16129: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6C).
Method: electron tomography / : Klein S, Chlanda P

EMDB-16130: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6E)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16131: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6G)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16132: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6I,K,M)
Method: electron tomography / : Klein S, Chlanda P

EMDB-16133: 
Tomogram of a late endosome of A549 cell infected with influenza A virus (Figure 6O)
Method: electron tomography / : Klein S, Chlanda P

EMDB-28616: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

PDB-8eun: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
