-Search query
-Search result
Showing 1 - 50 of 1,891 items for (author: fu & x)

EMDB-44486: 
Structure of apo-state V. cholerae DdmD
Method: single particle / : Shen ZF, Yang XY, Fu TM

EMDB-45254: 
Structure of V. cholerae monomeric DdmD bound with ssDNA
Method: single particle / : Shen ZF, Yang XY, Fu TM

EMDB-38389: 
Cryo-EM structure of sheep VMAT2 dimer in an atypical fold
Method: single particle / : Lyu Y, Fu C, Ma H, Sun Z, Su Z, Zhou X

EMDB-38390: 
Cryo-EM structure of a frog VMAT2 in an apo conformation
Method: single particle / : Lyu Y, Fu C, Ma H, Sun Z, Su Z, Zhou X

PDB-8xit: 
Cryo-EM structure of sheep VMAT2 dimer in an atypical fold
Method: single particle / : Lyu Y, Fu C, Ma H, Sun Z, Su Z, Zhou X

PDB-8xiu: 
Cryo-EM structure of a frog VMAT2 in an apo conformation
Method: single particle / : Lyu Y, Fu C, Ma H, Sun Z, Su Z, Zhou X

EMDB-41770: 
Apo form of human ATE1
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-42071: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

PDB-8uau: 
human ATE1 in complex with Arg-tRNA and a peptide substrate
Method: single particle / : Huang W, Zhang Y, Taylor DJ

EMDB-27922: 
Human TRPM2 ion channel in 1 mM ADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-27923: 
Human TRPM2 ion channel in 1 mM dADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-27924: 
Human TRPM2 ion channel in 1 mM dADPR and Ca2+
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-27925: 
Human TRPM2 ion channel in 1 mM BR-ADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-27926: 
Human TRPM2 ion channel in 1 mM F-dADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-27927: 
MHR1/2 and NUDT9H of human TRPM2 in 1 mM dADPR (local refinement)
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6q: 
Human TRPM2 ion channel in 1 mM ADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6r: 
Human TRPM2 ion channel in 1 mM dADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6s: 
Human TRPM2 ion channel in 1 mM dADPR and Ca2+
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6t: 
Human TRPM2 ion channel in 1 mM BR-ADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6u: 
Human TRPM2 ion channel in 1 mM F-dADPR
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

PDB-8e6v: 
MHR1/2 and NUDT9H of human TRPM2 in 1 mM dADPR (local refinement)
Method: single particle / : Wang L, Fu TM, Xia S, Wu H

EMDB-43128: 
Structure of the human dopamine transporter in complex with beta-CFT, MRS7292 and divalent zinc
Method: single particle / : Srivastava DK, Gouaux E

EMDB-36914: 
Cryo-EM structure of Streptomyces coelicolor transcription initiation complex with the global transcription factor AfsR
Method: single particle / : Lin W, Shi J, Xu JC
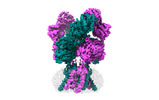
EMDB-43779: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation
Method: single particle / : Chou TH, Furukawa H
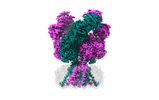
EMDB-43780: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in nonactive1 conformation
Method: single particle / : Chou TH, Furukawa H
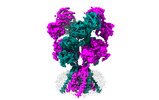
EMDB-43781: 
Rat GluN1-GluN2B NMDA receptor channel in apo conformation
Method: single particle / : Chou TH, Furukawa H
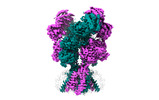
EMDB-43782: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine
Method: single particle / : Chou TH, Furukawa H

EMDB-43783: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate
Method: single particle / : Chou TH, Furukawa H
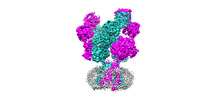
EMDB-44586: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry
Method: single particle / : Chou TH, Furukawa H

PDB-9are: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation
Method: single particle / : Chou TH, Furukawa H

PDB-9arf: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in nonactive1 conformation
Method: single particle / : Chou TH, Furukawa H

PDB-9arg: 
Rat GluN1-GluN2B NMDA receptor channel in apo conformation
Method: single particle / : Chou TH, Furukawa H

PDB-9arh: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine
Method: single particle / : Chou TH, Furukawa H

PDB-9ari: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glutamate
Method: single particle / : Chou TH, Furukawa H

PDB-9bib: 
Rat GluN1-GluN2B NMDA receptor channel in complex with glycine, glutamate, and EU-1622-A, in open-channel conformation, C1 symmetry
Method: single particle / : Chou TH, Furukawa H

EMDB-39110: 
Type I-EHNH Cascade complex
Method: single particle / : Li Z

EMDB-39167: 
Type I-FHNH Cascade-dsDNA intermediate complex
Method: single particle / : Li Z

EMDB-39200: 
Type I-FHNH Cascade-dsDNA R-loop complex
Method: single particle / : Li Z

EMDB-39285: 
Type I-FHNH Cascade complex
Method: single particle / : Li Z

EMDB-39286: 
Type I-EHNH Cascade-ssDNA complex
Method: single particle / : Li Z
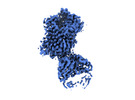
EMDB-19978: 
Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor
Method: single particle / : Pedersen CN, Yang F, Ita S, Xu Y, Akunuri R, Trampari S, Neumann CMT, Desdorf LM, Schioett B, Salvino JM, Mortensen OV, Nissen P, Shahsavar A
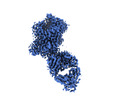
EMDB-19979: 
Inhibitor-free outward-open structure of Drosophila dopamine transporter
Method: single particle / : Pedersen CN, Yang F, Ita S, Xu Y, Akunuri R, Trampari S, Neumann CMT, Desdorf LM, Schioett B, Salvino JM, Mortensen OV, Nissen P, Shahsavar A

PDB-9euo: 
Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor
Method: single particle / : Pedersen CN, Yang F, Ita S, Xu Y, Akunuri R, Trampari S, Neumann CMT, Desdorf LM, Schioett B, Salvino JM, Mortensen OV, Nissen P, Shahsavar A

PDB-9eup: 
Inhibitor-free outward-open structure of Drosophila dopamine transporter
Method: single particle / : Pedersen CN, Yang F, Ita S, Xu Y, Akunuri R, Trampari S, Neumann CMT, Desdorf LM, Schioett B, Salvino JM, Mortensen OV, Nissen P, Shahsavar A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search









 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
