-Search query
-Search result
Showing 1 - 50 of 3,443 items for (author: fu & c)

EMDB entry, No image
EMDB-41434: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB entry, No image
EMDB-41435: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB entry, No image
EMDB-41436: 
Central rod disk in D3 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB entry, No image
EMDB-41463: 
Synechocystis PCC 6803 Phycobilisome quenched by OCP, high resolution
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB entry, No image
EMDB-41475: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB entry, No image
EMDB-41585: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to2: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to5: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tpj: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tro: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB entry, No image
EMDB-42520: 
CNQX-bound GluN1a-3A NMDA receptor
Method: single particle / : Michalski K, Furukawa H

EMDB entry, No image
EMDB-42522: 
Glycine-bound GluN1a-3A NMDA receptor
Method: single particle / : Michalski K, Furukawa H

EMDB entry, No image
EMDB-42580: 
Glycine-bound GluN1a-3A LBD heterotetramer (local refinement)
Method: single particle / : Michalski K, Furukawa H

PDB-8usw: 
CNQX-bound GluN1a-3A NMDA receptor
Method: single particle / : Michalski K, Furukawa H

PDB-8usx: 
Glycine-bound GluN1a-3A NMDA receptor
Method: single particle / : Michalski K, Furukawa H

PDB-8uue: 
Glycine-bound GluN1a-3A LBD heterotetramer (local refinement)
Method: single particle / : Michalski K, Furukawa H

EMDB entry, No image
EMDB-41963: 
Preholo-Proteasome from Beta 3 D205 deletion
Method: single particle / : Walsh Jr RM, Rawson S, Velez B, Blickling M, Razi A, Hanna J

EMDB entry, No image
EMDB-41993: 
Proteasome 20S Core Particle from Beta 3 D205 deletion
Method: single particle / : Walsh Jr RM, Rawson S, Velez B, Blickling M, Razi A, Hanna J

EMDB-15697: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Fuglsang A, Pape T, Mesa P, Stutzke LS, Temperini P, Montoya G

PDB-8axa: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
Method: single particle / : Tenjo-Castano F, Sofos N, Stella S, Fuglsang A, Pape T, Mesa P, Stutzke LS, Temperini P, Montoya G

EMDB-19406: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-19407: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8rox: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8roy: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L
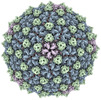
EMDB-40271: 
Mycobacterium phage Adjutor
Method: single particle / : Podgorski JM, White SJ

EMDB-37850: 
Cryo-EM structure of native H. thermoluteolus TH-1 GroEL
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37853: 
Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37862: 
Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-37863: 
Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wu4: 
Cryo-EM structure of native H. thermoluteolus TH-1 GroEL
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wuc: 
Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wuw: 
Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

PDB-8wux: 
Cryo-EM structure of H. thermophilus GroEL-GroES bullet complex
Method: single particle / : Liao Z, Gopalasingam CC, Kameya M, Gerle C, Shigematsu H, Ishii M, Arakawa T, Fushinobu S

EMDB-36870: 
Structure of full Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36871: 
In situ structure of RNA-dependent RNA polymerase in full BAV particles
Method: single particle / : Li Z, Cao S

EMDB-36872: 
Structure of VP9 in Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36880: 
Structure of partial Banna virus
Method: single particle / : Li Z, Cao S

EMDB-36881: 
Structure of Banna virus core
Method: single particle / : Li Z, Cao S

EMDB-37378: 
Structure of full Banna virus
Method: single particle / : Li Z, Cao S

EMDB-37379: 
Structure of partial Banna virus
Method: single particle / : Li Z, Cao S

EMDB-37380: 
Structure of Banna virus core
Method: single particle / : Li Z, Cao S

PDB-8k43: 
In situ structure of RNA-dependent RNA polymerase in full BAV particles
Method: single particle / : Li Z, Cao S

EMDB-17171: 
CTE typeI tau filament from Guam ALS/PDC
Method: helical / : Qi C, Yang S, Scheres SHW, Goedert M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search










 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
