-Search query
-Search result
Showing 1 - 50 of 119 items for (author: fraser & p)

EMDB-17297: 
Structure of a human 48S translation initiation complex with eIF4F and eIF4A
Method: single particle / : Brito Querido J, Sokabe M, Diaz-Lopez I, Gordiyenko Y, Fraser CS, Ramakrishnan V

PDB-8oz0: 
Structure of a human 48S translation initiation complex with eIF4F and eIF4A
Method: single particle / : Brito Querido J, Sokabe M, Diaz-Lopez I, Gordiyenko Y, Fraser CS, Ramakrishnan V
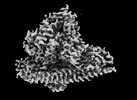
EMDB-41617: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M, Sun M

PDB-8tu6: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M

EMDB-27876: 
E. coli 50S ribosome bound to tiamulin and VS1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27877: 
E. coli 50S ribosome bound to tiamulin and azithromycin
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e41: 
E. coli 50S ribosome bound to tiamulin and VS1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e42: 
E. coli 50S ribosome bound to tiamulin and azithromycin
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27878: 
E. coli 50S ribosome bound to compound streptogramin A analog 3336
Method: single particle / : Pellegrino J, Lee DJ, Seiple IB, Fraser JS

PDB-8e43: 
E. coli 50S ribosome bound to compound streptogramin A analog 3336
Method: single particle / : Pellegrino J, Lee DJ, Seiple IB, Fraser JS

EMDB-27867: 
E. coli 50S ribosome bound to D-linker solithromycin conjugate
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27868: 
E. coli 50S ribosome bound to L-linker solithromycin conjugate
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27869: 
E. coli 50S ribosome bound to solithromycin and VM1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e3l: 
E. coli 50S ribosome bound to D-linker solithromycin conjugate
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e3m: 
E. coli 50S ribosome bound to L-linker solithromycin conjugate
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e3o: 
E. coli 50S ribosome bound to solithromycin and VM1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27852: 
E. coli 50S ribosome bound to compound streptogramin A analog 3142
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27854: 
E. coli 50S ribosome bound to compound streptogramin analogs SA1 and SB1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27855: 
E. coli 50S ribosome bound to compound streptogramin analog SAB001
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27857: 
E. coli 50S ribosome bound to compound SAB002
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27858: 
E. coli 50S ribosome bound to compound streptogramin A analog 3146
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e30: 
E. coli 50S ribosome bound to compound streptogramin A analog 3142
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e32: 
E. coli 50S ribosome bound to compound streptogramin analogs SA1 and SB1
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e33: 
E. coli 50S ribosome bound to compound streptogramin analog SAB001
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e35: 
E. coli 50S ribosome bound to compound SAB002
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e36: 
E. coli 50S ribosome bound to compound streptogramin A analog 3146
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27879: 
E. coli 50S ribosome bound to antibiotic analog SLC09
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27880: 
E. coli 50S ribosome bound to antibiotic analog SLC17
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27881: 
E. coli 50S ribosome bound to antibiotic analog SLC21
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27882: 
E. coli 50S ribosome bound to antibiotic analog SLC26
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27883: 
E. coli 50S ribosome bound to antibiotic analog SLC30
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-27884: 
E. coli 50S ribosome bound to antibiotic analog SLC31
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e44: 
E. coli 50S ribosome bound to antibiotic analog SLC09
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e45: 
E. coli 50S ribosome bound to antibiotic analog SLC17
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e46: 
E. coli 50S ribosome bound to antibiotic analog SLC21
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e47: 
E. coli 50S ribosome bound to antibiotic analog SLC26
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e48: 
E. coli 50S ribosome bound to antibiotic analog SLC30
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

PDB-8e49: 
E. coli 50S ribosome bound to antibiotic analog SLC31
Method: single particle / : Pellegrino J, Lee DJ, Fraser JS, Seiple IB

EMDB-16330: 
Botulinum neurotoxin serotype X in complex with NTNH/X
Method: single particle / : Martinez-Carranza M, Skerlova J, Stenmark P

PDB-8byp: 
Botulinum neurotoxin serotype X in complex with NTNH/X
Method: single particle / : Martinez-Carranza M, Skerlova J, Stenmark P

EMDB-25443: 
CryoEM map of the manually-picked stalled contraction intermediate state of bacteriophage A511.
Method: single particle / : Fraser A, Leiman PG

EMDB-25444: 
CryoEM map of the CNN-picked stalled contraction intermediate state of bacteriophage A511.
Method: single particle / : Fraser A, Leiman PG

EMDB-24763: 
Cryo-EM map of the phage AR9 non-virion RNA polymerase holoenzyme in complex with DNA containing the AR9 P077 promoter
Method: single particle / : Fraser A, Leiman PG, Sokolova ML
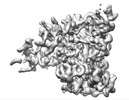
EMDB-24765: 
Cryo-EM map of the phage AR9 non-virion RNA polymerase holoenzyme
Method: single particle / : Fraser A, Leiman PG, Sokolova ML

PDB-7um0: 
Structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with two DNA oligonucleotides containing the AR9 P077 promoter as determined by cryo-EM
Method: single particle / : Leiman PG, Fraser A, Sokolova ML

PDB-7um1: 
Structure of bacteriophage AR9 non-virion RNAP polymerase holoenzyme determined by cryo-EM
Method: single particle / : Leiman PG, Fraser A, Sokolova ML

EMDB-26657: 
50S E. coli ribosome bound to streptogramins VM2 and VS1
Method: single particle / : Pellegrino J, Lee DJ

EMDB-26067: 
CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B.
Method: single particle / : Lapointe CP, Grosely R, Sokabe M, Alvarado C, Wang J, Montabana E, Villa N, Shin B, Dever T, Fraser C, Fernandez IS, Puglisi JD

PDB-7tql: 
CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B.
Method: single particle / : Lapointe CP, Grosely R, Sokabe M, Alvarado C, Wang J, Montabana E, Villa N, Shin B, Dever T, Fraser C, Fernandez IS, Puglisi JD

EMDB-24802: 
Wild-type Escherichia coli stalled ribosome with antibiotic radezolid
Method: single particle / : Young ID, Stojkovic V, Tsai K, Lee DJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
