-Search query
-Search result
Showing all 25 items for (author: fernandez-tornero & c)

EMDB-19492: 
Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase
Method: single particle / : Ruiz FM, Plaza-Pegueroles A, Fernandez-Tornero C

PDB-8rth: 
Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase
Method: single particle / : Ruiz FM, Plaza-Pegueroles A, Fernandez-Tornero C

EMDB-14421: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40).
Method: single particle / : Nguyen PQ, Huecas S, Plaza-Pegueroles A, Fernandez-Tornero C

EMDB-14468: 
Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A
Method: single particle / : Nguyen PQ, Fernandez-Tornero C

EMDB-14469: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.9 A (focus subunit C11 terminal Zn-ribbon in the funnel pore).
Method: single particle / : Nguyen PQ, Huecas S, Plaza-Pegueroles A, Fernandez-Tornero C

EMDB-14470: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore).
Method: single particle / : Nguyen PQ, Huecas S, Plaza-Pegueroles A, Fernandez-Tornero C

EMDB-16299: 
Structure of yeast RNA Polymerase III elongation complex at 3.3 A
Method: single particle / : Nguyen PQ, Fernandez-Tornero C

PDB-7z0h: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40).
Method: single particle / : Nguyen PQ, Huecas S, Plaza-Pegueroles A, Fernandez-Tornero C

PDB-7z2z: 
Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A
Method: single particle / : Nguyen PQ, Fernandez-Tornero C

PDB-7z30: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.9 A (focus subunit C11 terminal Zn-ribbon in the funnel pore).
Method: single particle / : Nguyen PQ, Fernandez-Tornero C

PDB-7z31: 
Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore).
Method: single particle / : Nguyen PQ, Huecas S, Plaza-Pegueroles A, Fernandez-Tornero C

PDB-8bws: 
Structure of yeast RNA Polymerase III elongation complex at 3.3 A
Method: single particle / : Nguyen PQ, Fernandez-Tornero C

EMDB-0146: 
Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD)
Method: single particle / : Sanz-Murillo M, Xu J, Gil-Carton D, Wang D, Fernandez-Tornero C

EMDB-0147: 
Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49
Method: single particle / : Sanz-Murillo M, Xu J, Gil-Carton D, Wang D, Fernandez-Tornero C

PDB-6h67: 
Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD)
Method: single particle / : Sanz-Murillo M, Xu J, Gil-Carton D, Wang D, Fernandez-Tornero C

PDB-6h68: 
Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49
Method: single particle / : Sanz-Murillo M, Xu J, Gil-Carton D, Wang D, Fernandez-Tornero C

EMDB-4086: 
RNA polymerase I-Rrn3 complex
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

EMDB-4087: 
Monomeric RNA polymerase I at 5.6 A resolution
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

EMDB-4088: 
Monomeric RNA polymerase I at 4.9 A resolution
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C

PDB-5lmx: 
Monomeric RNA polymerase I at 4.9 A resolution
Method: single particle / : Torreira E, Louro JA, Gil-Carton D, Gallego O, Calvo O, Fernandez-Tornero C
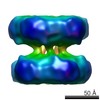
EMDB-2581: 
The structures of cytosolic and plastid-located glutamine synthetases from Medicago truncatula reveal a common and dynamic architecture
Method: single particle / : Torreira E, Seabra AR, Marriott H, Zhou M, Llorca O, Robinson CV, Carvalho HG, Fernandez-Tornero C, Pereira PJB

EMDB-1802: 
Conformational flexibility of RNA polymerase III during transcriptional elongation
Method: single particle / : Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Muller CW

EMDB-1803: 
Conformational flexibility of RNA polymerase III during transcriptional elongation
Method: single particle / : Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Muller CW

EMDB-1804: 
Conformational flexibility of RNA polymerase III during transcriptional elongation
Method: single particle / : Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Muller CW

EMDB-1322: 
Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III.
Method: single particle / : Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Sentenac A, Muller CW, Schoehn G
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
