-Search query
-Search result
Showing all 48 items for (author: fernandez & jj)

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
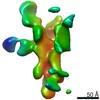
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
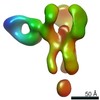
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
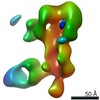
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
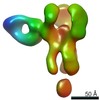
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25646: 
Negative stain map of polyclonal Fab 241.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25655: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA
Method: single particle / : Han J, Ward AB

EMDB-24364: 
A-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24366: 
B-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24367: 
Partial of A-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24368: 
A-tubule of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24370: 
B-tubule of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24371: 
A-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24372: 
B-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24373: 
Partial A-tubule of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24374: 
A-tubule of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24375: 
B-tubule of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24376: 
Inner junction structure of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24377: 
An unidentified MIP at the inner junction of axonemal microtubule doublet from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24379: 
Composite map of axonemal microtubule doublet in 16 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24380: 
Composite map of axonemal microtubule doublet in 48 nm repeat from Tetrahymena thermophila
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24381: 
Composite map of axonemal microtubule doublet in 16 nm repeat from a Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-24382: 
Composite map of axonemal microtubule doublet in 48 nm repeat from a Tetrahymena thermophila mutant fap115 knockout
Method: subtomogram averaging / : Li S, Fernandez JJ, Fabritius A, Agard DA, Winey M

EMDB-13183: 
Subtomogram average of T20S proteasome from Thermoplasma acidophilum
Method: subtomogram averaging / : Fernandez JJ, Li S

EMDB-13184: 
Subtomogram average of Axonemal Doublet Microtubule from Tetrahymena thermophila
Method: subtomogram averaging / : Fernandez JJ, Li S

EMDB-12285: 
Platelet integrin from intact cells
Method: subtomogram averaging / : Sorrentino S, Conesa JJ, Cuervo A, Melero R, Martins B, Fernandez-Gimenez E, Isidro-Gomez F, Jimenez de la Morena J, Studt JD, Sorzano COS, Eibauer M, Carazo JM, Medalia O

EMDB-12531: 
T20S proteasome subtomogram average
Method: subtomogram averaging / : Fernandez JJ

EMDB-4667: 
Cryo-EM structure of T7 bacteriophage portal protein, 13mer, closed valve
Method: single particle / : Cuervo A, Fabrega-Ferrer M, Machon C, Fernandez FJ, Perez-Luque R, Pous J, Vega MC, Carrascosa JL, Coll M

EMDB-4669: 
Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve
Method: single particle / : Fabrega-Ferrer M, Cuervo A, Machon C, Fernandez FJ, Perez-Luque R, Pous J, Vega MC, Carrascosa JL, Coll M

EMDB-4706: 
Cryo-EM structure of T7 bacteriophage fiberless tail complex
Method: single particle / : Cuervo A, Fabrega-Ferrer M, Machon C, Conesa JJ, Perez-Ruiz M, Coll M, Carrascosa JL

EMDB-9167: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9168: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9169: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9170: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9171: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9172: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9173: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-9174: 
Electron cryo-tomography and subtomogram averaging of microtubule triplet from procentriole
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall W, Agard DA

EMDB-2799: 
Cryo-EM structure of gamma-TuSC oligomers in a closed conformation
Method: helical / : Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong K, Fernandez JJ, Sali A, Kilmartin J, Davis TN, Agard DA

EMDB-5989: 
Electron cryo-tomography of the Microtubule Nucleation Complex in the Yeast Spindle Pole Body
Method: subtomogram averaging / : Kollman JM, Greenberg C, Li S, Fernandez JJ, Moritz M, Zelter A, Fong K, Sali A, Kilmartin JV, Davis TN, Agard DA

EMDB-5252: 
electron cryo-tomography reconstruction and subvolume averaging of the basal body triplet from Chlamydomonas reinhardtii
Method: subtomogram averaging / : Li S, Fernandez JJ, Marshall WF, Agard DA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
