-Search query
-Search result
Showing 1 - 50 of 440 items for (author: fang & pa)

EMDB-37104: 
96-nm axonemal repeat with RS1/2/3
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37111: 
48-nm repeat DMT
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37114: 
Radial Spoke 1 (RS1)
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37116: 
RS1 refined with head mask
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37117: 
Radial Spoke 2 (RS2)
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37118: 
Radial Spoke 2 (RS2) head
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37119: 
Radial Spoke 3
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37120: 
Radial Spoke 3 head
Method: subtomogram averaging / : Cong X, Yao C

EMDB-37516: 
BA.2(S375) Spike (S6P)/hACE2 complex
Method: single particle / : Wei X, Zhang Z

EMDB-37517: 
Local refinement of RBD-ACE2
Method: single particle / : Wei X, Zhang Z

EMDB-37949: 
RS head-neck monomer
Method: single particle / : Meng X, Cong Y

EMDB-38003: 
The consensus map of RS head-neck dimer
Method: single particle / : Meng X, Cong Y

EMDB-38004: 
consensus map of RS head-neck monomer complex
Method: single particle / : Meng X, Cong Y

EMDB-38013: 
Rsph16-Rsph16' bridge region of Radial spoke head-neck dimer
Method: single particle / : Meng X, Cong Y

EMDB-38014: 
Radial spoke head-neck dimer's monomer-half focuses-refinement map
Method: single particle / : Meng X, Cong Y

EMDB-38019: 
RS head-neck dimer's neck focused-refinement map
Method: single particle / : Meng X, Cong Y

EMDB-38020: 
Radial spoke head-neck dimer
Method: single particle / : Meng X, Cong Y

EMDB-38028: 
Radial spoke head-neck complex, focus-refinement on core
Method: single particle / : Meng X, Cong Y

EMDB-38029: 
Focus-refinement on RS head-neck monomer's arm 1
Method: single particle / : Meng X, Cong Y

EMDB-38030: 
Focus-refinement on RS head-neck monomer's arm 2
Method: single particle / : Meng X, Cong Y

EMDB-38031: 
Focus-refinement on RS head-neck monomer's neck
Method: single particle / : Meng X, Cong Y

EMDB-29794: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29795: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29796: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29797: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g72: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g73: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g74: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g75: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F
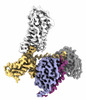
EMDB-35463: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y
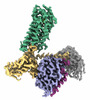
EMDB-35483: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y
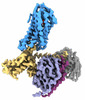
EMDB-35484: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y
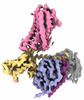
EMDB-35485: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y

PDB-8ij3: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y

PDB-8ija: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y

PDB-8ijb: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y

PDB-8ijd: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y

EMDB-35880: 
Human KCNQ2-CaM in complex with CBD and PIP2
Method: single particle / : Ma D, Li X, Guo J

EMDB-35881: 
Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II
Method: single particle / : Ma D, Li X, Guo J

EMDB-37270: 
human KCNQ2-CaM in complex with PIP2 and HN37
Method: single particle / : Ma D, Li X, Guo J

PDB-8j02: 
Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II
Method: single particle / : Ma D, Li X, Guo J

PDB-8w4u: 
human KCNQ2-CaM in complex with PIP2 and HN37
Method: single particle / : Ma D, Li X, Guo J

EMDB-35882: 
Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state I
Method: single particle / : Ma D, Li D, Guo J

EMDB-35883: 
Human KCNQ2-CaM-HN37 complex in the presence of PIP2
Method: single particle / : Ma D, Li X, Guo J

EMDB-35884: 
Human KCNQ2-CaM complex in the presence of PIP2
Method: single particle / : Ma D, Li X, Guo J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search








 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
