-Search query
-Search result
Showing 1 - 50 of 67 items for (author: efremov & rg)

EMDB-16157: 
Map of GroEL:GroES-(ADP) complex plunge frozen 200 ms after reaction initiation with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-40676: 
Human TRP channel TRPV6 in cNW30 nanodiscs inhibited by tetrahydrocannabivarin (THCV)
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

PDB-8sp8: 
Human TRP channel TRPV6 in cNW30 nanodiscs inhibited by tetrahydrocannabivarin (THCV)
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

EMDB-16091: 
Cryo-EM structure of beta-galactosidase at 3.3 A resolution plunged 5 ms after mixing with apoferritin
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16092: 
Cryo-EM structure of beta-galactosidase at 2.9 A resolution plunged 205 ms after mixing with apoferritin
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16093: 
Cryo-EM structure of mouse heavy-chain apoferritin at 2.1 A plunged 5ms after mixing with b-galactosidase
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16094: 
Cryo-EM structure of mouse heavy-chain apoferritin at 2.7 A plunged 35ms after mixing with b-galactosidase
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16095: 
Cryo-EM structure of mouse heavy-chain apoferritin at 2.2 A plunged 205ms after mixing with b-galactosidase
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16097: 
Cryo-EM structure of beta-galactosidase at 3.2 A resolution plunged 35 ms after mixing with apoferritin
Method: single particle / : Torino S, Dhurandhar M, Efremov R

EMDB-16099: 
GroEL:GroES-ATP complex under continuous turnover condirions
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16100: 
Structure of GroEL-ATP complex plunge frozen 200 ms after reaction initiation
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16102: 
Structure of GroEL-nucleotide complex in ADP-like conformation plunged 13 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16106: 
Structure of the GroEL-ATP complex plunge-frozen 50 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16107: 
Structure of the GroEL(ATP7/ADP7) complex plunged 13 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16108: 
Structure of GroEL-nucleotide complex in ADP-like conformation plunged 50 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16109: 
Structure of the GroEL(ATP7/ADP7) complex plunged 50 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16115: 
Structure of the GroEL-ATP complex plunge-frozen 13 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16116: 
Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16117: 
Structure of GroEL:GroES-ATP complex under continuous turnover conditions
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16118: 
Structure of GroEL-ATP complex under continuous turnover conditions
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16119: 
Structure of GroEL:GroES complex exhibiting ADP-conformation in trans ring obtained under the continuous turnover conditions
Method: single particle / : Dhurandhar M, Torino S, Efremov R

EMDB-16125: 
Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation
Method: single particle / : Dhurandhar M, Efremov R, Torino S

EMDB-16154: 
Map of the GroEL-ES-ATP complex plunge-frozen 50 ms after mixing with ATP
Method: single particle / : Dhurandhar M, Torino S, Efremov R
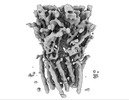
EMDB-16308: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo state (dataset 1)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
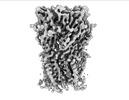
EMDB-16314: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo in apo state (dataset 2)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
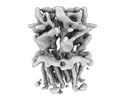
EMDB-16315: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo state (Alpo4_LMNG_Serotonin dataset 4)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
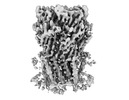
EMDB-16316: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo state (Alpo4_comb dataset 3)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
EMDB-16317: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in apo state (Alpo4_apo, dataset 1)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
EMDB-16326: 
Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in complex with CHAPS(Alpo4_CHAPS)
Method: single particle / : De Gieter S, Efremov RG, Ulens C
EMDB-29343: 
Cryo-EM structure of human TRPV6 in complex with the natural phytoestrogen genistein
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

EMDB-29344: 
Cryo-EM structure of human TRPV6 in the open state
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

PDB-8foa: 
Cryo-EM structure of human TRPV6 in complex with the natural phytoestrogen genistein
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

PDB-8fob: 
Cryo-EM structure of human TRPV6 in the open state
Method: single particle / : Neuberger A, Yelshanskaya MV, Nadezhdin KD, Sobolevsky AI

EMDB-15213: 
Cryo-EM density map of Tn4430 TnpA hyperactive mutant (TnpA3X) in complex with IR48 substrate.
Method: single particle / : Shkumatov AV, Liu Y, Efremov RG

EMDB-15218: 
Medium resolution cryo-EM density map of Tn4430 TnpA transposase from Tn3 family in apo state
Method: single particle / : Shkumatov AV, Liu Y, Efremov RG

EMDB-13906: 
Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA
Method: single particle / : Shkumatov AV, Oger CA, Aryanpour N, Hallet BF, Efremov RG

EMDB-13908: 
Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA
Method: single particle / : Shkumatov AV, Oger CA, Aryanpour N, Hallet BF, Efremov RG

EMDB-13909: 
Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product
Method: single particle / : Shkumatov AV, Oger CA, Aryanpour N, Hallet BF, Efremov RG

EMDB-13910: 
Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state
Method: single particle / : Shkumatov AV, Oger CA, Aryanpour N, Hallet BF, Efremov RG

EMDB-12653: 
Respiratory complex I from Escherichia coli - conformation 1
Method: single particle / : Kolata P, Efremov RG

EMDB-12654: 
Respiratory complex I from Escherichia coli - conformation 2
Method: single particle / : Kolata P, Efremov RG

EMDB-12655: 
Respiratory complex I from Escherichia coli - conformation 3
Method: single particle / : Kolata P, Efremov RG

EMDB-13291: 
Respiratory complex I from Escherichia coli solubilized in LMNG
Method: single particle / : Kolata P, Efremov RG

EMDB-12652: 
Respiratory complex I from Escherichia coli - focused refinement of membrane arm
Method: single particle / : Kolata P, Efremov RG

EMDB-12661: 
Respiratory complex I from Escherichia coli - focused refinement of cytoplasmic arm
Method: single particle / : Kolata P, Efremov RG

EMDB-23853: 
Mouse TRPV3 in MSP2N2 nanodiscs, closed state at 4 degrees Celsius
Method: single particle / : Neuberger A, Nadezhdin KD, Sobolevsky AI

EMDB-23854: 
Mouse TRPV3 in MSP2N2 nanodiscs, closed state at 42 degrees Celsius
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-23855: 
Mouse TRPV3 in MSP2N2 nanodiscs, sensitized state at 42 degrees Celsius
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI

EMDB-23856: 
Mouse TRPV3 in cNW11 nanodiscs, closed state at 4 degrees Celsius
Method: single particle / : Neuberger A, Nadezhdin KD, Sobolevsky AI

EMDB-23857: 
Mouse TRPV3 in cNW11 nanodiscs, closed state at 42 degrees Celsius
Method: single particle / : Nadezhdin KD, Neuberger A, Sobolevsky AI
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
