-Search query
-Search result
Showing 1 - 50 of 187 items for (author: dun & se)
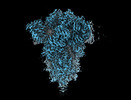
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
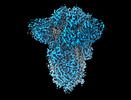
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1c: 
SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1d: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

PDB-8cin: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-29675: 
A Vibrio cholerae viral satellite enables efficient horizontal transfer by using an external scaffold to assemble hijacked coat proteins into small capsids
Method: single particle / : Subramanian S, Boyd CM, Seed KD, Parent KN

PDB-8g1r: 
A Vibrio cholerae viral satellite enables efficient horizontal transfer by using an external scaffold to assemble hijacked coat proteins into small capsids
Method: single particle / : Subramanian S, Boyd CM, Seed KD, Parent KN

EMDB-17391: 
Subtomogram average of ribosomes in germinated polar tubes of Vairimorpha necatrix
Method: subtomogram averaging / : Sharma H, Ehrenbolger K, Jespersen N, Carlson LA, Barandun J

EMDB-17467: 
Subtomogram representing a segment of the outer wall of empty germinated polar tubes from Vairimorpha necatrix.
Method: subtomogram averaging / : Sharma H, Ehrenbolger K, Jespersen N, Carlson LA, Barandun J

EMDB-17468: 
Subtomogram representing a segment of the outer wall of cargo-filled, germinated polar tubes from Vairimorpha necatrix.
Method: subtomogram averaging / : Sharma H, Ehrenbolger K, Jespersen N, Carlson LA, Barandun J

EMDB-41370: 
Structure of a class A GPCR/Fab complex
Method: single particle / : Sun D, Johnson M, Masureel M

EMDB-41827: 
Structure of a class A GPCR/agonist complex (focused map2)
Method: single particle / : Sun D, Johnson M, Masureel M

EMDB-41828: 
Structure of a class A GPCR/agonist complex (focused map1)
Method: single particle / : Sun D, Johnson M, Masureel M

EMDB-41829: 
Structure of a class A GPCR/agonist complex
Method: single particle / : Sun D, Johnson M, Masureel M

EMDB-41850: 
Structure of a class A GPCR/agonist complex (Consensus map)
Method: single particle / : Sun D, Johnson M, Masureel M

PDB-8tlm: 
Structure of a class A GPCR/Fab complex
Method: single particle / : Sun D, Johnson M, Masureel M

PDB-8u1u: 
Structure of a class A GPCR/agonist complex
Method: single particle / : Sun D, Johnson M, Masureel M

EMDB-29783: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

EMDB-41613: 
Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074
Method: single particle / : Tolbert WD, Pozharski E, Pazgier M

PDB-8g6u: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

PDB-8ttw: 
Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074
Method: single particle / : Tolbert WD, Pozharski E, Pazgier M

EMDB-27596: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

PDB-8dok: 
Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074
Method: single particle / : Chen Y, Zhou F, Huang R, Tolbert W, Pazgier M

EMDB-15311: 
Dedicated chaperone at the ribosome safeguards the proteostasis network during eEF1A biogenesis
Method: single particle / : Minoia M, Quintana-Cordero J, Katharina J, Kotan I, Turnbull KJ, Masser AE, Merker D, Gouarin E, Ehrenbolger K, Barandun J, Hauryliuk V, Bukau B, Kramer G, Andreasson C

EMDB-27593: 
The structure of S. epidermidis Cas10-Csm bound to target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA
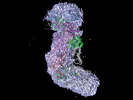
EMDB-27762: 
318 kDa Cas10-Csm effector complex bound to cognate target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA

PDB-8do6: 
The structure of S. epidermidis Cas10-Csm bound to target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA

EMDB-35326: 
Cryo-EM reconstruction of PBCV-1 capsid block 1
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35490: 
Cryo-EM reconstruction of PBCV-1 capsid block 2
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35585: 
Cryo-EM reconstruction of PBCV-1 capsid block 3
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35586: 
Cryo-EM reconstruction of PBCV-1 capsid block 4
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35589: 
Cryo-EM reconstruction of PBCV-1 capsid block 5
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35593: 
Cryo-EM reconstruction of PBCV-1 capsid block 6
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35594: 
Cryo-EM reconstruction of PBCV-1 capsid block 8
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35644: 
Cryo-EM reconstruction of PBCV-1 capsid block 9
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35645: 
Cryo-EM reconstruction of PBCV-1 capsid block 7
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35646: 
Cryo-EM reconstruction of PBCV-1 capsid block 10
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35647: 
Cryo-EM reconstruction of PBCV-1 capsid block 11
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35648: 
Cryo-EM reconstruction of PBCV-1 capsid block 12
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35650: 
Cryo-EM reconstruction of PBCV-1 capsid block 13
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35656: 
Cryo-EM reconstruction of PBCV-1 capsid block 14
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35659: 
Cryo-EM reconstruction of PBCV-1 capsid block 15
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35695: 
Cryo-EM reconstruction of PBCV-1 capsid block 16
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35954: 
Cryo-EM reconstruction of PBCV-1 capsid block 17
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35955: 
Cryo-EM reconstruction of PBCV-1 capsid block 18
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35956: 
Cryo-EM reconstruction of PBCV-1 capsid block 19
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35957: 
Cryo-EM reconstruction of PBCV-1 capsid block 20
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35958: 
Cryo-EM reconstruction of PBCV-1 capsid block 21
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35959: 
Cryo-EM reconstruction of PBCV-1 capsid block 22
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
