-Search query
-Search result
Showing 1 - 50 of 78 items for (author: dong & zy)

EMDB-34929: 
Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H

EMDB-34927: 
Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H

EMDB-33625: 
Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-33626: 
Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM

EMDB-33627: 
Cryo-EM structure of the left-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-33630: 
Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM
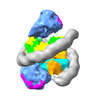
EMDB-33631: 
Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-35660: 
Composite cryo-EM density map of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM
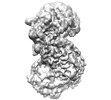
EMDB-35661: 
Cryo-EM structure of the monomeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35708: 
Cryo-EM consensus map of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35709: 
Local refinement cryo-EM map of protomer I of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35710: 
Local refinement cryo-EM map of protomer II of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-36013: 
Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-36014: 
Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-33474: 
Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1
Method: single particle / : Yang G, Xu L, Chang S, Guo J, Wu Z

EMDB-33472: 
Structure of ATP7B C983S/C985S/D1027A mutant in presence of ATOX1
Method: single particle / : Yang G, Xu L, Guo J, Wu Z

EMDB-33475: 
Structure of ATP7B C983S/C985S/D1027A mutant
Method: single particle / : Yang G, Xu L, Guo J, Wu Z

EMDB-33476: 
Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1
Method: single particle / : Yang G, Xu L, Chang S, Guo J, Wu Z

EMDB-35628: 
Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP
Method: single particle / : Yang G, Xu L, Guo J, Wu Z

EMDB-31968: 
Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1
Method: single particle / : Wang YD, Wang LX, Guan ZY, Yin P

EMDB-24346: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-80 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24348: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-081 Fab
Method: single particle / : Saphire EO, Li H

EMDB-24349: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-091 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24350: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-094 VHH-Fc
Method: single particle / : Saphire EO, Li H

EMDB-24351: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-096 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24358: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-147 IgG
Method: single particle / : Saphire EO, Yu X

EMDB-24359: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-246 IgG
Method: single particle / : Saphire EO, Rayaprolu V

EMDB-24335: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-259 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24336: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-186 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24337: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-199 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24338: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-249 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24339: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-252 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24340: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-049 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24341: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-073 scFv
Method: single particle / : Saphire EO, Li H

EMDB-24342: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-043 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24343: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-010 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24344: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-148 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24345: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-002 Fab
Method: single particle / : Saphire EO, Li H

EMDB-24352: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-250 Fab
Method: single particle / : Saphire EO, Li H

EMDB-24353: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-063 scFv
Method: single particle / : Saphire EO, Li H

EMDB-24354: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-021 Fab
Method: single particle / : Saphire EO, Li H

EMDB-24355: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-247 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24356: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-020 Fab
Method: single particle / : Saphire EO, Li H

EMDB-24357: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-090 IgG
Method: single particle / : Saphire EO, Hui S

EMDB-24360: 
EM map of SARS-CoV-2 Spike in complex with CoVIC-245 IgG
Method: single particle / : Saphire EO, Rayaprolu V

EMDB-24361: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-074 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24383: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-140 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24384: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-134 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24388: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-038 IgG
Method: single particle / : Saphire EO, Li H

EMDB-24293: 
Negative stain EM map of SARS-CoV-2 Spike in complex with human ACE2
Method: single particle / : Saphire EO, Li H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
