-Search query
-Search result
Showing 1 - 50 of 674 items for (author: das & s)

EMDB-43889: 
Chlamydomonas reinhardtii mastigoneme (constituent map 1)
Method: single particle / : Dai J, Ma M, Zhang R, Brown A

EMDB-43890: 
Chlamydomonas reinhardtii mastigoneme (constituent map 2)
Method: single particle / : Dai J, Ma M, Zhang R, Brown A

EMDB-43891: 
Chlamydomonas reinhardtii mastigoneme (constituent map 3)
Method: single particle / : Dai J, Ma M, Zhang R, Brown A

EMDB-43892: 
Composite cryo-EM map of the Chlamydomonas reinhardtii mastigoneme
Method: single particle / : Dai J, Ma M, Zhang R, Brown A

PDB-9b4h: 
Chlamydomonas reinhardtii mastigoneme filament
Method: single particle / : Dai J, Ma M, Zhang R, Brown A

EMDB-17329: 
48S late-stage initiation complex with m6A mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

EMDB-17330: 
48S late-stage initiation complex with non methylated mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

PDB-8p03: 
48S late-stage initiation complex with m6A mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y

PDB-8p09: 
48S late-stage initiation complex with non methylated mRNA
Method: single particle / : Guca E, Lima LHF, Boissier F, Hashem Y
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1c: 
SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1d: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-42820: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5b extended
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

PDB-8cin: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-17993: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

PDB-8pwh: 
Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex
Method: single particle / : Ruedas R, Vuillemot R, Tubiana T, Winter JM, Pieri L, Arteni AA, Samson C, Jonic J, Mathieu M, Bressanelli S

EMDB-17757: 
Cryo-EM structure of the Cas12m-crRNA-target DNA complex
Method: single particle / : Sasnauskas G, Tamulaitiene G, Karvelis T, Bigelyte G, Siksnys V

PDB-8pm4: 
Cryo-EM structure of the Cas12m-crRNA-target DNA complex
Method: single particle / : Sasnauskas G, Tamulaitiene G, Karvelis T, Bigelyte G, Siksnys V

EMDB-15411: 
Single particle structure of Atg18-WT
Method: single particle / : Mann D, Fromm S, Martinez-Sanchez A, Gopaldass N, Mayer A, Sachse C

PDB-8afx: 
Single particle structure of Atg18-WT
Method: single particle / : Mann D, Fromm S, Martinez-Sanchez A, Gopaldass N, Mayer A, Sachse C

PDB-8pvv: 
Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 30 nt RNA guide and 51 nt DNA target
Method: single particle / : Manakova EN, Zaremba M, Pocevicuite R, Golovinas E, Sasnauskas G, Zagorskaite E, Silanskas A

PDB-8qg0: 
Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target
Method: single particle / : Manakova EN, Zaremba M, Pocevicuite R, Golovinas E, Zagorskaite E, Silanskas A

EMDB-18043: 
Helical structure of the influenza A virus ribonucleoprotein-like
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T
EMDB-18044: 
Focused reconstruction of influenza A RNP-like particle
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

PDB-8pzp: 
Model for influenza A virus helical ribonucleoprotein-like structure
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

PDB-8pzq: 
Model for focused reconstruction of influenza A RNP-like particle
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

EMDB-42816: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyp: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-27141: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2)
Method: single particle / : Rai J, Das A, Li H
EMDB-27142: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1)
Method: single particle / : Rai J, Das A, Li H
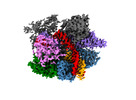
EMDB-27143: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage)
Method: single particle / : Rai J, Das A, Li H
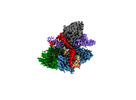
EMDB-27144: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2)
Method: single particle / : Rai J, Das A, Li H
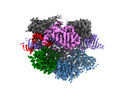
EMDB-27145: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound)
Method: single particle / : Rai J, Das A, Li H
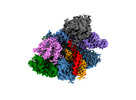
EMDB-27146: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2k: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2l: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 1)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2n: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2o: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2p: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Target bound)
Method: single particle / : Rai J, Das A, Li H

PDB-8d2q: 
Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1)
Method: single particle / : Rai J, Das A, Li H

EMDB-42801: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42802: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42805: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42808: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 4
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42809: 
MERS 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42810: 
MERS 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
