-Search query
-Search result
Showing 1 - 50 of 126 items for (author: chang & yc)

EMDB-36150: 
Cryo-EM structure of Vibrio campbellii alpha-hemolysin
Method: single particle / : Wang CH, Yeh MK, Ho MC, Lin SM

EMDB-35598: 
cryo-EM structure of the middle part of the shrimp white spot syndrome virus nucleocapsid (wide type)
Method: single particle / : Huang HJ, Wang HC, Chen LL

EMDB-35600: 
Cryo-Em structure of the middle part of the shrimp white spot syndrome virus nucleocapsid (narrow type)
Method: single particle / : Huang HJ, Wang HC

EMDB-33145: 
Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form
Method: single particle / : Wang CH, Hsieh JT, Ho MC, Hung HC

EMDB-33146: 
Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA
Method: single particle / : Wang CH, Hsieh JT, Ho MC, Hung HC

EMDB-33147: 
Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor MDSA
Method: single particle / : Wang CH, Hsieh JT, Ho MC, Hung HC

EMDB-31820: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD
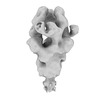
EMDB-31818: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Gamma variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31822: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Kappa variant in complex with neutralizing antibody RBD-chAb-25
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31821: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibody RBD-chAb-25
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31817: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Beta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD

EMDB-31819: 
Negative staining (NS)-EM structure of SARS-CoV-2 S-Delta variant in complex with neutralizing antibodies, RBD-chAb-15 and RBD-chAb45
Method: single particle / : Yu PY, Yang TJ, Chang YC, Wu HC, Hsu STD
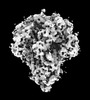
EMDB-32329: 
Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32332: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32333: 
Subtomogram averaging of PEDV (Pintung 52) S protein with one protomer in the D0-up conformation and two protomers in the D0-down conformation, determined in situ on intact viral particles
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32337: 
Subtomogram averaging of PEDV (Pintung 52) S protein with two protomers in the D0-up conformation and one protomer in the D0-down conformation, determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32338: 
Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
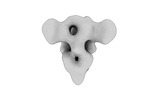
EMDB-32339: 
Subtomogram averaging of PEDV (Pintung 52) S protein with all three protomers in the D0-up conformation determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-32340: 
Subtomogram averaging of PEDV (Pintung 52) S protein in the postfusion form determined in situ on intact viral particles.
Method: subtomogram averaging / : Hsu STD, Draczkowski P, Wang YS, Huang CY

EMDB-33646: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33647: 
Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33648: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
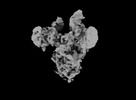
EMDB-33649: 
Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33700: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33701: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33702: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S protein with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33703: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-33704: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-up and two D0-down
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33705: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I one D0-down and two D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS
EMDB-33706: 
Cryo-EM map of HEK293F cell-derived PEDV PT52 S T326I with three D0-up
Method: single particle / : Hsu STD, Draczkowski P, Wang YS

EMDB-32832: 
SARS-CoV-2 Spike in complex with Fab of m31A7
Method: single particle / : Wu YM, Chen X

EMDB-32825: 
Negative stain volume of the mono-GlcNAc-decorated SARS-CoV-2 Spike
Method: single particle / : Chen X, Huang HY

EMDB-25448: 
Negative-stain EM reconstruction of SpFN_1B-06-PL, a SARS-CoV-2 spike fused to H.pylori ferritin nanoparticle vaccine candidate
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25449: 
RFN_131, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Receptor-Binding Domain
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25450: 
pCoV146, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike Receptor-Binding and N-Terminal Domains
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-25451: 
pCoV111, a Ferritin-based Nanoparticle Vaccine Candidate Displaying the SARS-CoV-2 Spike S1 Subunit
Method: single particle / : Thomas PV, Smith C, Chen WH, Sankhala RS, Hajduczki A, Choe M, Martinez E, Chang W, Peterson CE, Karch C, Gohain N, Kannadka CB, de Val N, Joyce MG, Modjarrad K

EMDB-31760: 
Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, one RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31761: 
Cryo-EM structure of SARS-CoV-2 S-Beta variant (B.1.351), uncleavable form, two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31762: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31763: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), one RBD-up conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31764: 
Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1), two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31767: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), all RBD-down conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31768: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31769: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), one RBD-up conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31770: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), two RBD-up conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31771: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31772: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 2
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31773: 
Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1), dimer of S trimer conformation 3
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31775: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), all RBD-down conformation
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD

EMDB-31776: 
Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2), one RBD-up conformation 1
Method: single particle / : Yang TJ, Yu PY, Chang YC, Hsu STD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
