-Search query
-Search result
Showing 1 - 50 of 71 items for (author: burns & e)

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
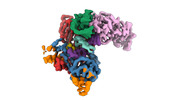
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
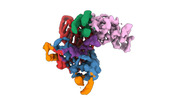
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-27249: 
Yeast mitochondrial small subunit assembly intermediate (State 1)
Method: single particle / : Burnside C, Harper N, Klinge S

EMDB-27250: 
Yeast mitochondrial small subunit assembly intermediate (State 2)
Method: single particle / : Burnside C, Harper NJ, Klinge S

PDB-8d8j: 
Yeast mitochondrial small subunit assembly intermediate (State 1)
Method: single particle / : Burnside C, Harper N, Klinge S

PDB-8d8k: 
Yeast mitochondrial small subunit assembly intermediate (State 2)
Method: single particle / : Burnside C, Harper NJ, Klinge S

EMDB-26966: 
Human mitochondrial small subunit assembly intermediate (State A)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-26967: 
Human mitochondrial small subunit assembly intermediate (State B)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-26968: 
Human mitochondrial small subunit assembly intermediate (State C)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-26969: 
Human mitochondrial small subunit assembly intermediate (State D)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-26970: 
Human mitochondrial small subunit assembly intermediate (State E)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-26971: 
Human mitochondrial small subunit assembly intermediate (State C*)
Method: single particle / : Harper NJ, Burnside C, Klinge S

EMDB-27251: 
Yeast mitochondrial small subunit assembly intermediate (State 3)
Method: single particle / : Burnside C, Harper N, Klinge S

PDB-8csp: 
Human mitochondrial small subunit assembly intermediate (State A)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8csq: 
Human mitochondrial small subunit assembly intermediate (State B)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8csr: 
Human mitochondrial small subunit assembly intermediate (State C)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8css: 
Human mitochondrial small subunit assembly intermediate (State D)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8cst: 
Human mitochondrial small subunit assembly intermediate (State E)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8csu: 
Human mitochondrial small subunit assembly intermediate (State C*)
Method: single particle / : Harper NJ, Burnside C, Klinge S

PDB-8d8l: 
Yeast mitochondrial small subunit assembly intermediate (State 3)
Method: single particle / : Burnside C, Harper N, Klinge S

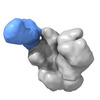
EMDB-25621: 
The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex with 3 copies of the neutralizing antibody K11
Method: single particle / : Berndsen ZT, Ward AB
EMDB-25623: 
SIVmac239.K180S SOSIP in complex with FZ019.2 Fab
Method: single particle / : Berndsen ZT

EMDB-25624: 
SIVmac239.K180S SOSIP in complex with FZ012.7 Fab
Method: single particle / : Berndsen ZT, Lee W

EMDB-25625: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque Rh33519
Method: single particle / : Berndsen ZT, Lee W

EMDB-25626: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque Rh31186
Method: single particle / : Berndsen ZT, Lee W

EMDB-25627: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque Rh34620
Method: single particle / : Berndsen ZT, Lee W

EMDB-25628: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque r11008
Method: single particle / : Berndsen ZT, Lee W

EMDB-25629: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque r11008
Method: single particle / : Berndsen ZT, Lee W

EMDB-25630: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque r11002
Method: single particle / : Berndsen ZT, Lee W

EMDB-25631: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque r11004
Method: single particle / : Berndsen ZT, Lee W

EMDB-25632: 
SIVmac239.K180S SOSIP in complex with an unknown Fab from the polyclonal serum of the SIV-infected Rhesus Macaque r11004
Method: single particle / : Berndsen ZT, Lee W
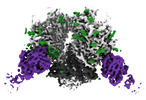
EMDB-25676: 
The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex with 3 copies of the neutralizing antibody K11
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t2p: 
The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex with 3 copies of the neutralizing antibody K11
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t4g: 
The Envelope Glycoprotein SIVmac239.K180S SOSIP trimer in complex with 3 copies of the neutralizing antibody K11
Method: single particle / : Berndsen ZT, Ward AB

EMDB-24693: 
SARS-CoV-2-6P-Mut7 S protein (C3 symmetry)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24694: 
SARS-CoV-2-6P-Mut7 S protein (asymmetric)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24695: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24696: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24697: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

EMDB-24699: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru1: 
SARS-CoV-2-6P-Mut7 S protein (C3 symmetry)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru2: 
SARS-CoV-2-6P-Mut7 S protein (asymmetric)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru3: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru4: 
CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru5: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB

PDB-7ru8: 
CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement)
Method: single particle / : Ozorowski G, Turner HL, Ward AB
EMDB-26467: 
SARS-CoV-2 6P Mut7 in complex with Fab THSC20.HVTR04 (1 RBD up and 1 RDB down)
Method: single particle / : Torres JL, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
