-Search query
-Search result
Showing all 37 items for (author: bujnicki & j)

EMDB-18520: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18521: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18522: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

EMDB-18523: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo2: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM
PDB-8qo3: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo4: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM

PDB-8qo5: 
Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes
Method: single particle / : Moura TR, Purta E, Bernat A, Baulin E, Mukherjee S, Bujnicki JM
EMDB-34047: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Ling X, Yang X, Wu Y, Liu T, Wei X, Bujnick JM, Zhao K, Su Z
EMDB-34048: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Liu L, Ling X, Yang X, Wu Y, Liu T, Miao Z, Wei X, Bujnicki JM, Zhao K, Su Z

PDB-7yr6: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Ling X, Yang X, Wu Y, Liu T, Wei X, Bujnick JM, Zhao K, Su Z

PDB-7yr7: 
Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers
Method: single particle / : Jia X, Pan Z, Yuan Y, Luo B, Luo Y, Mukherjee S, Jia G, Liu L, Ling X, Yang X, Wu Y, Liu T, Miao Z, Wei X, Bujnicki JM, Zhao K, Su Z
EMDB-33136: 
The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-33137: 
The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd5: 
The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd6: 
The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-33134: 
The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

EMDB-33135: 
The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

EMDB-33138: 
The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-34670: 
The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
EMDB-34671: 
The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z
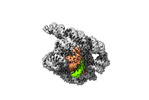
EMDB-35223: 
The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd3: 
The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd4: 
The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-7xd7: 
The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-8hd6: 
The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-8hd7: 
The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

PDB-8i7n: 
The Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside
Method: single particle / : Luo B, Zhang C, Ling X, Mukherjee S, Jia G, Xie J, Jia X, Liu L, Baulin EF, Luo Y, Jiang L, Dong H, Wei X, Bujnicki JM, Su Z

EMDB-4958: 
Negative stain EM 3D reconstruction of the UvrA-UvrB-DNA complex.
Method: single particle / : Swuec P, Renault L, Costa A

EMDB-3232: 
The architecture of the S. pombe CCR4-NOT complex
Method: single particle / : Ukleja M, Cuellar J, Siwaszek A, Kasprzak JM, Czarnocki-Cieciura M, Bujnicki J, Dziembowski A, Valpuesta JM
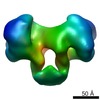
EMDB-1890: 
EcoR124 Type I DNA restriction-modification enzyme complex in closed state with bound 30bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1891: 
EcoR124 Type I DNA restriction-modification enzyme complex without DNA (open state). Low resolution 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
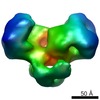
EMDB-1892: 
EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1893: 
EcoKI Type I DNA restriction-modification enzyme complex in closed state with bound 75bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

PDB-2y7c: 
Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534.
Method: single particle / : Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF

PDB-2y7h: 
Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534.
Method: single particle / : Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF

EMDB-1534: 
EcoKI type I RM methyltransferase with DNA mimic Ocr. Negative stain 3D.
Method: single particle / : Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
