-Search query
-Search result
Showing 1 - 50 of 52 items for (author: bubeck & d)

EMDB-18742: 
Structure of interleukin 6.
Method: single particle / : Gardner S, Bubeck D, Jin Y

EMDB-18743: 
Structure of interleukin 6 (gp130 P496L mutant).
Method: single particle / : Gardner S, Bubeck D, Jin Y

PDB-8qy6: 
Structure of interleukin 6 (gp130 P496L mutant).
Method: single particle / : Gardner S, Bubeck D, Jin Y

EMDB-18741: 
Structure of interleukin 11 (gp130 P496L mutant).
Method: single particle / : Gardner S, Bubeck D, Jin Y

PDB-8qy4: 
Structure of interleukin 11 (gp130 P496L mutant).
Method: single particle / : Gardner S, Bubeck D, Jin Y
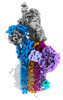
EMDB-15779: 
CryoEM structure of C5b8-CD59
Method: single particle / : Bubeck D, Couves EC, Gardner S

EMDB-15780: 
2C9, C5b9-CD59 structure
Method: single particle / : Couves EC, Gardner S, Bubeck D
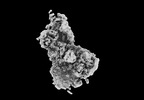
EMDB-15781: 
2C9, C5b9-CD59 cryoEM structure
Method: single particle / : Couves EC, Gardner S, Bubeck D

EMDB-15782: 
2C9, C5b9-CD59 cryoEM structure; focus refinement map
Method: single particle / : Couves EC, Gardner S, Bubeck D
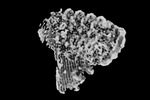
EMDB-15783: 
3C9, C5b9-CD59 cryoEM structure; focus refinement map
Method: single particle / : Couves EC, Gardner S, Bubeck D
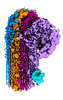
EMDB-15800: 
CryoEM reconstruction of C5b8-CD59, density subtracted
Method: single particle / : Bubeck D, Couves EC, Gardner S

EMDB-14427: 
IL-27 signalling complex
Method: single particle / : Jin Y, Gardner S, Bubeck D

EMDB-12646: 
cryoEM reconstruction of the terminal C9s and chaperone in 3C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-12647: 
cryoEM reconstruction of the terminal C9s in 2C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-12648: 
cryoEM reconstruction of C5b and C7 C-ter in 2C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-12649: 
cryoEM reconstruction of 1C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-12650: 
cryoEM structure of 3C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-12651: 
cryoEM structure of 2C9-sMAC
Method: single particle / : Menny A, Couves EC, Bubeck D

EMDB-11172: 
Disulfide-locked early prepore intermedilysin-CD59
Method: single particle / : Shah NR, Bubeck D

PDB-6zd0: 
Disulfide-locked early prepore intermedilysin-CD59
Method: single particle / : Shah NR, Bubeck D

EMDB-0071: 
GAPDH-CP12-PRK complex
Method: single particle / : McFarlane CR, Shah N, Bubeck D, Murray JW

PDB-6gve: 
GAPDH-CP12-PRK complex
Method: single particle / : McFarlane CR, Shah N, Bubeck D, Murray JW

EMDB-0106: 
OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX
Method: single particle / : Menny A, Serna M, Boyd CM, Gardner S, Joseph AP, Topf M, Bubeck D

EMDB-0107: 
Closed conformation of the Membrane Attack Complex
Method: single particle / : Menny A, Serna M, Boyd CM, Gardner S, Joseph AP, Topf M, Bubeck D

EMDB-0109: 
Asymmetric region of the Membrane Attack Complex in the closed conformation
Method: single particle / : Menny A, Serna M, Boyd CB, Gardener S, Joseph AP, Topf M, Bubeck D

EMDB-0110: 
Consensus map of the Membrane Attack Complex
Method: single particle / : Menny A, Serna M, Boyd CB, Gardener S, Joseph AP, Topf M, Bubeck D

EMDB-0111: 
Asymmetric region of the Membrane Attack Complex in the open conformation
Method: single particle / : Menny A, Serna M, Boyd CB, Gardener S, Joseph AP, Topf M, Bubeck D

EMDB-0112: 
Hinge region of the Membrane Attack Complex in the open conformation
Method: single particle / : Menny A, Serna M, Boyd CB, Gardener S, Joseph AP, Topf M, Bubeck D

EMDB-0113: 
C9 oligomer region of the Membrane Attack Complex in the open conformation
Method: single particle / : Menny A, Serna M, Boyd CB, Gardener S, Joseph AP, Topf M, Bubeck D

PDB-6h03: 
OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX
Method: single particle / : Menny A, Serna M, Boyd CM, Gardner S, Joseph AP, Topf M, Bubeck D

PDB-6h04: 
Closed conformation of the Membrane Attack Complex
Method: single particle / : Menny A, Serna M, Boyd CM, Gardner S, Joseph AP, Topf M, Bubeck D
EMDB-1991: 
Structure of complement component complex, sC5b9.
Method: single particle / : Bubeck D, Roversi P, Hakabyan S, Morgan BP, Lea SM, Llorca O
EMDB-1964: 
Structural and Functional Studies of LRP6 Ectodomain Reveal a Platform for Wnt Signaling
Method: single particle / : Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY
EMDB-1965: 
Structural and Functional Studies of LRP6 Ectodomain Reveal a Platform for Wnt Signaling
Method: single particle / : Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY

EMDB-1805: 
Structure of human complement C8, a precursor to membrane attack.
Method: single particle / : Bubeck D, Roversi P, Donev R, Morgan BP, Llorca O, Lea SM

EMDB-1206: 
Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging.
Method: single particle / : Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ

EMDB-1207: 
Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging.
Method: single particle / : Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ

EMDB-1144: 
The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
Method: single particle / : Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM

EMDB-1145: 
The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
Method: single particle / : Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM

PDB-1xyr: 
Poliovirus 135S cell entry intermediate
Method: single particle / : Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM

EMDB-1136: 
The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
Method: single particle / : Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search












 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
