-Search query
-Search result
Showing 1 - 50 of 231 items for (author: briggs & jag)
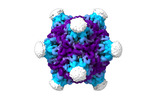
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
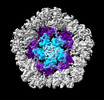
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
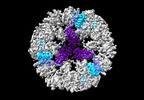
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
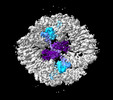
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-16698: 
Subtomogram average of HIV-1 CA pentamer from capsid-like particles assembled with inositol hexakisphosphate
Method: subtomogram averaging / : Tan A, Briggs JAG, Dick RA

EMDB-16699: 
Subtomogram average of HIV-1 CA hexamer from capsid-like particles assembled with inositol hexakisphosphate
Method: subtomogram averaging / : Tan A, Briggs JAG, Dick RA

EMDB-29772: 
HIV-1 CA lattice bound to IP6; from capsid-like particles
Method: single particle / : Highland CM, Dick RA

EMDB-29773: 
HIV-1 capsid lattice bound to IP6, pH 6.2
Method: single particle / : Highland CM, Dick RA

EMDB-29774: 
HIV-1 CA lattice bound to IP6, pH 7.4
Method: single particle / : Highland CM, Dick RA

EMDB-29775: 
HIV-1 capsid lattice bound to dNTPs
Method: single particle / : Highland CM, Dick RA

EMDB-29776: 
HIV-1 capsid lattice bound to IP6 and Lenacapavir
Method: single particle / : Highland CM, Dick RA

EMDB-29777: 
Defective HIV-1 CA pentamer in surrounding hexameric lattice
Method: single particle / : Highland CM, Dick RA

EMDB-16703: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16704: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16705: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16706: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16707: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16708: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs bound to Nup153 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16709: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to CPSF6 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16710: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to CPSF6 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16711: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Sec24C peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16712: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Sec24C peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16183: 
In situ structure of the Caulobacter crescentus S-layer
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16207: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.2 A from 5 tomograms
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

EMDB-16209: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.0 A from the full dataset
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

EMDB-15949: 
COPII inner coat reprocessed with relion4.0
Method: subtomogram averaging / : Zanetti G, Pyle E

EMDB-33748: 
Spike_GSAS_6P protomer RBD domain bound with R1-32 Fab and ACE2 with 3:3:3 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
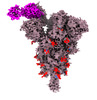
EMDB-33760: 
Spike_GSAS_6P and R1-32 Fab with 3to1 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
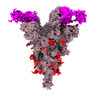
EMDB-33764: 
SARS-COV-2 Spike_GSAS_6P bound with two R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
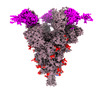
EMDB-33766: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

EMDB-33772: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

EMDB-33453: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33454: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33455: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33456: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33457: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33458: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33459: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33460: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33461: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site) after 40-Day Storage in PBS, Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33462: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33463: 
Structure of SARS-CoV-2 D614G spike protein (single Arg S1/S2 cleavage site), Closed Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-33464: 
Structure of SARS-Cov-2 D614G spike protein (single Arg S1/S2 cleavage site), Open Conformation
Method: single particle / : Qu K, Chen Q, Ciazynska KA, Liu B, Zhang X, Wang J, He Y, Guan J, He J, Liu T, Carter AP, Xiong X, Briggs JAG

EMDB-14517: 
Chimera of AP2 with FCHO2 linker domain as a fusion on Cmu2 subunit
Method: single particle / : Kane Dickson V, Qu K, Owen DJ, Briggs JA, Zaccai NR

EMDB-14518: 
Chimaera of AP2 with FCHO2 linker domain, N1-N2 enriched population
Method: single particle / : Kane Dickson V, Qu K, Owen DJ, Briggs JA, Zaccai NR

EMDB-14525: 
AP2 adaptor protein recruited on the membrane in the presence of FCHO2 linker
Method: electron tomography / : Kovtun O, Kaufman JGG, Owen DJ, Briggs JAG

EMDB-14526: 
AP2 on the membrane without cargo peptide
Method: subtomogram averaging / : Kovtun O, Kaufman JGG, Owen DJ, Briggs JAG

EMDB-13085: 
Representative cryo-electron tomogram of immature HIV-1 particles
Method: electron tomography / : Qu K, Ke ZL, Zila V, Anders-Oesswein M, Glass B, Muecksch F, Mueller R, Schultz C, Mueller B, Kraeusslich HG, Briggs JAG

EMDB-13086: 
Representative cryo-electron tomogram of mature HIV-1 particles
Method: electron tomography / : Qu K, Ke ZL, Zila V, Anders-Oesswein M, Glass B, Muecksch F, Mueller R, Schultz C, Mueller B, Kraeusslich HG, Briggs JAG

EMDB-13087: 
Immature HIV-1 matrix structure
Method: subtomogram averaging / : Qu K, Ke ZL, Zila V, Anders-Oesswein M, Glass B, Muecksch F, Mueller R, Schultz C, Mueller B, Kraeusslich HG, Briggs JAG
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
