-Search query
-Search result
Showing 1 - 50 of 145 items for (author: braun & n)
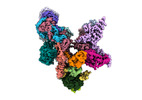
EMDB-18496: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18499: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map A
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18500: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map B.
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18502: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map D
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18503: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba.- Map E
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18504: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map F
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

PDB-8qma: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS
EMDB-18501: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map C
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-16158: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 peripheral arm)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16159: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 CIII)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16160: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 membrane arm)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16161: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 peripheral arm)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16163: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 CIII)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-17597: 
cryo-EM structure of Doa10 in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

EMDB-17608: 
cryo-EM structure of Doa10 with RING domain in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

EMDB-17609: 
Low resolution map of Doa10 in MSP1E3D1
Method: single particle / : Botsch JJ, Schulman BA

EMDB-17610: 
Doa10 in MSP2N2
Method: single particle / : Botsch JJ, Schulman BA

EMDB-16172: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bq6: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16171: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 1 composition)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bq5: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 1 composition)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16156: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 membrane arm)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16168: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bpx: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16153: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete consensus)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16166: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 2 consensus)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16167: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Conformation 1 consensus)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-15998: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI peripheral tip)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-15999: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI peripheral core)
Method: single particle / : Klusch N, Kuehlbrandt W
EMDB-16000: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16003: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16007: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-16008: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII MPP domain)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bed: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI peripheral tip)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bee: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI peripheral core)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bef: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8beh: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bel: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain)
Method: single particle / : Klusch N, Kuehlbrandt W

PDB-8bep: 
Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII MPP domain)
Method: single particle / : Klusch N, Kuehlbrandt W

EMDB-26121: 
Skd3_ATPyS_FITC-casein Hexamer, AAA+ only
Method: single particle / : Rizo AN

EMDB-26122: 
Skd3, hexamer, filtered
Method: single particle / : Rizo AN, Cupo RR

EMDB-12707: 
O-Layer C14 at 2.58A - Local refinement with C14 symmetry of the O-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.
Method: single particle / : Mace K, Vadakkepat AK, Lukoyanova N, Waksman G

EMDB-12708: 
I-Layer C16 at 3.08A - Local refinement with C16 symmetry of I-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.
Method: single particle / : Mace K, Vadakkepat AK, Lukoyanova N, Waksman G

EMDB-12709: 
Stalk C1 at 3.71A - Local refinement without symmetry of the Stalk complex from the fully-assembled R388 type IV secretion system.
Method: single particle / : Mace K, Vadakkepat AK, Lukoyanova N, Waksman G

EMDB-12715: 
IMC-Arches C6 at 8.33A - Refinement with C6 symmetry of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system.
Method: single particle / : Mace K, Vadakkepat AK, Lukoyanova N, Waksman G
EMDB-12716: 
Trimer of dimers TrwK/VirB4unbound C1 at 4.14A - Refinement without symmetry of TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system.
Method: single particle / : Vadakkepat AK, Mace K, Lukoyanova N, Waksman G

EMDB-12717: 
Dimer TrwK/VirB4unbound C1 at 3.49A - Local refinement without symmetry of the TrwK/VirB4unbound dimer complex from the R388 type IV secretion system.
Method: single particle / : Vadakkepat AK, Mace K, Lukoyanova N, Waksman G

EMDB-12933: 
IMC protomer C1 at 3.75A - Local refinement without symmetry of the inner membrane complex (IMC) protomer from the fully-assembled R388 type IV secretion system.
Method: single particle / : Mace K, Vadakkepat AK, Lukoyanova N, Waksman G
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
