-Search query
-Search result
Showing 1 - 50 of 196 items for (author: borgnia & mj)

EMDB-41730: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin
Method: single particle / : Wright NJ, Lee SY
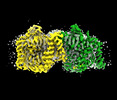
EMDB-41731: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in MSP2N2 nanodiscs, apo state
Method: single particle / : Wright NJ, Lee SY
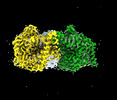
EMDB-41732: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY
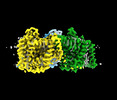
EMDB-41733: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction
Method: single particle / : Wright NJ, Lee SY
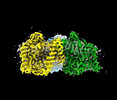
EMDB-41734: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with N-hydroxycytidine
Method: single particle / : Wright NJ, Lee SY
EMDB-41735: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with PSI-6206
Method: single particle / : Wright NJ, Lee SY

EMDB-41736: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41737: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41738: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT2 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41739: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41740: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY

EMDB-41741: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-OFS conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41742: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41743: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41744: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-OFS conformation (from ensemble analysis)
Method: single particle / : Wright NJ, Lee SY
EMDB-41745: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-OFS conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41746: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT1 conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41747: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41748: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (clockwise) conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41749: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (counterclockwise) conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41750: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41751: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT1-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41752: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY
EMDB-41755: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-OFS conformation (3DVA analysis)
Method: single particle / : Wright NJ, Lee SY

EMDB-29172: 
Cryo-EM structure of Cryptococcus neoformans trehalose-6-phosphate synthase homotetramer in complex with uridine diphosphate and glucose-6-phosphate
Method: single particle / : Washington EJ, Brennan RG

EMDB-29426: 
Chaetomium thermophilum SETX - NPPC internal deletion
Method: single particle / : Williams RS, Appel CD

EMDB-29439: 
Chaetomium thermophilum SETX (Full-length)
Method: single particle / : Williams RS, Appel CD

EMDB-29440: 
SETX-Sen1N domain
Method: single particle / : Williams RS, Appel CD

EMDB-27889: 
Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class II, C1 state with endogenous PI(4,5)P2 bound
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27890: 
Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class III, putative desensitized state
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27701: 
Focused map (monomer A) for Arabidopsis SPY in complex with GDP-fucose
Method: single particle / : Kumar S, Zhou Y, Dillard L, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-27702: 
Focused map (monomer B) for Arabidopsis SPY in complex with GDP-fucose
Method: single particle / : Kumar S, Zhou Y, Lucas D, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-28586: 
Cryo-EM structure of the organic cation transporter 1 in the apo state
Method: single particle / : Suo Y, Wright NJ, Lee SY

EMDB-28587: 
Cryo-EM structure of the organic cation transporter 1 in complex with diphenhydramine
Method: single particle / : Suo Y, Wright NJ, Lee SY

EMDB-28588: 
Cryo-EM structure of the organic cation transporter 1 in complex with verapamil
Method: single particle / : Suo Y, Wright NJ, Lee SY

EMDB-28589: 
Cryo-EM structure of the organic cation transporter 2 in complex with 1-methyl-4-phenylpyridinium
Method: single particle / : Suo Y, Wright NJ, Lee SY

EMDB-28755: 
Human tRNA Splicing Endonuclease Complex bound to 2'F-tRNA-Arg
Method: single particle / : Stanley RE, Hayne CK

EMDB-27696: 
Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Method: single particle / : Kumar S, Zhou Y, Dillard L, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-27697: 
Cryo-EM structure of Arabidopsis SPY alternative conformation 1
Method: single particle / : Kumar S, Zhou Y, Dillard L, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-27698: 
Cryo-EM structure of Arabidopsis SPY alternative conformation 2
Method: single particle / : Kumar S, Zhou Y, Dillard L, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-27699: 
Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose
Method: single particle / : Kumar S, Zhou Y, Dillard L, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-27700: 
Overall map of Arabidopsis SPY in complex with GDP-fucose
Method: single particle / : Kumar S, Zhou Y, Lucas D, Borgnia MJ, Bartesaghi A, Zhou P

EMDB-26856: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG
Method: single particle / : Stanley RE, Hayne CK

EMDB-26831: 
Human Rix1 sub-complex scaffold
Method: single particle / : Gordon J, Stanley RE

EMDB-27891: 
The open state mouse TRPM8 structure in complex with the cooling agonist C3, AITC, and PI(4,5)P2
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27892: 
The intermediate C2-state mouse TRPM8 structure in complex with the cooling agonist C3 and PI(4,5)P2
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27893: 
The closed C1-state mouse TRPM8 structure in complex with PI(4,5)P2
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27894: 
The closed C1-state mouse TRPM8 structure in complex with putative PI(4,5)P2
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27895: 
Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class I , C0 state
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY

EMDB-27896: 
The closed C0-state flycatcher TRPM8 structure in complex with PI(4,5)P2
Method: single particle / : Yin Y, Zhang F, Feng S, Butay KJ, Borgnia MJ, Im W, Lee SY
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
