-Search query
-Search result
Showing 1 - 50 of 57 items for (author: black & ss)

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D
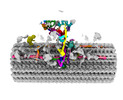
EMDB-41284: 
Combined linker domain of N-DRC and associated proteins Tetrahymena
Method: single particle / : Ghanaeian AG, Majhi SM, McCaffrey CM, Nami BN, Black CB, Yang SK, Legal TL, Papoulas OP, Janowska MJ, Valente-Paterno MV, Marcotte EM, Wloga DW, Bui KH

PDB-8tid: 
Combined linker domain of N-DRC and associated proteins Tetrahymena
Method: single particle / : Ghanaeian AG, Majhi SM, McCaffrey CM, Nami BN, Black CB, Yang SK, Legal TL, Papoulas OP, Janowska MJ, Valente-Paterno MV, Marcotte EM, Wloga DW, Bui KH

EMDB-41189: 
Baseplate of Nexin-dynein regulatory complex from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Black CS, Yang SK, Bui KH
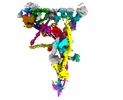
EMDB-41251: 
Linker domain of Nexin-dynein regulatory complex from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Bui KH

EMDB-41270: 
Focused refinement of the N-DRC from the Tetrahymena WT subtomo
Method: subtomogram averaging / : Ghanaeian AG, Majhi SM, McCaffrey CM, Nami BN, Black CB, Yang SK, Legal TL, Papoulas OP, Janowska MJ, Valente-Paterno MV, Marcotte EM, Wloga DW, Bui KH

EMDB-41375: 
Linker domain of N-DRC complex from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Bui KH

EMDB-41376: 
96nm repeat of Doublet microtubule from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Bui KH

EMDB-41504: 
DRC9/10 baseplate from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Bui KH

PDB-8tek: 
Baseplate of Nexin-dynein regulatory complex from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Black CS, Yang SK, Bui KH

PDB-8th8: 
Linker domain of Nexin-dynein regulatory complex from Tetrahymena thermophila
Method: single particle / : Ghanaeian AG, Bui KH

EMDB-40891: 
Main central pair structure
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40892: 
Ciliary cap complex from Tetrahymena thermophila
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40894: 
Singlet A-tubule from Tetrahymena thermophila
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40896: 
Tip central pair consensus map from Tetrahymena thermophila cilia
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40897: 
C1 refinement of the tip central pair subtomogram average
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40898: 
C1 MAPS refinement of ciliary tip central pair subtomogram average
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40899: 
C2 refinement of ciliary tip central pair subtomogram average
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40900: 
C2 MAPs refinement of ciliary tip central pair subtomogram average
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40909: 
Ciliary tip central pair map of FAP256-KO Tetrahymena thermophila (consensus map)
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40910: 
Ciliary tip central pair map of FAP256-KO Tetrahymena thermophila (C1 refinement)
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40911: 
Ciliary tip central pair map of FAP256-KO Tetrahymena thermophila (C2 refinement)
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40912: 
Ciliary tip central pair map of FAP256-KO Tetrahymena thermophila (composite map)
Method: subtomogram averaging / : Legal T, Bui KH

EMDB-40913: 
Ciliary tip tomogram of Tetrahymena thermophila
Method: electron tomography / : Legal T, Bui KH

EMDB-40299: 
Consensus refinement of the h12-LOX in a dimeric form
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40300: 
The local refinement map of a "closed" subunit of a 12-LOX dimer
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40301: 
The local refinement map of an "open" subunit of a 12-LOX dimer
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40302: 
The consensus map of a 12-LOX hexamer
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40304: 
The local refinement map of a single subunit of a 12-LOX hexamer
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40039: 
The structure of h12-LOX in monomeric form
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40040: 
The structure of h12-LOX in dimeric form
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40041: 
The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-40042: 
The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

PDB-8ghb: 
The structure of h12-LOX in monomeric form
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

PDB-8ghc: 
The structure of h12-LOX in dimeric form
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

PDB-8ghd: 
The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

PDB-8ghe: 
The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA
Method: single particle / : Black KA, Mobbs JI, Venugopal H, Thal DM, Glukhova A

EMDB-16847: 
CryoEM structure of holo e4D2
Method: single particle / : Yadav KNS, Hutchins G, Berger Schaffitzel C, Anderson R

EMDB-29666: 
96-nm repeat unit of intact doublet microtubule from Tetrahymena thermophila CFAP77AB-KO mutant
Method: subtomogram averaging / : Black CS, Legal T, Bui KH

EMDB-29667: 
96-nm repeat unit of intact doublet microtubule from Tetrahymena thermophila strain CU428
Method: subtomogram averaging / : Black CS, Legal T, Bui KH

EMDB-29685: 
48-nm doublet microtubule from Tetrahymena thermophila strain CU428
Method: single particle / : Black CS, Kubo S, Yang SK, Bui KH

EMDB-29692: 
48-nm doublet microtubule from Tetrahymena thermophila strain K40R
Method: single particle / : Black CS, Kubo S, Yang SK, Bui KH

EMDB-29693: 
96-nm doublet microtubule from combined Tetrahymena thermophila strains CU428 and K40R
Method: single particle / : Black CS, Kubo S, Yang SK, Bui KH

PDB-8g2z: 
48-nm doublet microtubule from Tetrahymena thermophila strain CU428
Method: single particle / : Black CS, Kubo S, Yang SK, Bui KH

PDB-8g3d: 
48-nm doublet microtubule from Tetrahymena thermophila strain K40R
Method: single particle / : Black CS, Kubo S, Yang SK, Bui KH

EMDB-29684: 
Cryo-EM structure of ADGRF1 coupled to miniGs/q
Method: single particle / : Jones D, Rawson S, Blacklow S

PDB-8g2y: 
Cryo-EM structure of ADGRF1 coupled to miniGs/q
Method: single particle / : Jones D, Rawson S, Blacklow S

EMDB-29044: 
Structure of Zanidatamab bound to HER2
Method: single particle / : Worrall LJ, Atkinson CE, Sanches M, Dixit S, Strynadka NCJ

EMDB-23926: 
The structure of the Tetrahymena thermophila outer dynein arm on doublet microtubule
Method: single particle / : Kubo S, Yang SK, Ichikawa M, Bui KH

PDB-7moq: 
The structure of the Tetrahymena thermophila outer dynein arm on doublet microtubule
Method: single particle / : Kubo S, Yang SK, Ichikawa M, Bui KH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
