-Search query
-Search result
Showing 1 - 50 of 176 items for (author: bjorkman & p)

EMDB-29581: 
HIV-1 BG505 SOSIP-HT2 in complex with CD4 (class II)
Method: single particle / : Dam KA, Fan C

EMDB-29582: 
HIV-1 BG505 SOSIP-HT2 in complex with CD4 (class III)
Method: single particle / : Dam KA, Fan C

EMDB-29583: 
HIV-1 BG505 SOSIP-HT1 in complex with CD4 and 17b Fab
Method: single particle / : Dam KA, Fan C

EMDB-29584: 
HIV-1 BG505 SOSIP-HT2 in complex with CD4 and 17b Fab
Method: single particle / : Dam KA, Fan C

EMDB-29601: 
HIV-1 B41 SOSIP-HT2 in complex with CD4
Method: single particle / : Dam KA, Fan C

EMDB-40437: 
Structure of HIV-1 BG505 SOSIP-HT1 in complex with CD4 (class II)
Method: single particle / : Dam KA, Fan C

EMDB-40438: 
Structure of HIV-1 BG505 SOSIP-HT1 (class III)
Method: single particle / : Dam KA, Fan C

EMDB-29579: 
Structure of HIV-1 BG505 SOSIP-HT1 in complex with one CD4 molecule
Method: single particle / : Fan C, Dam KA, Bjorkman PJ

EMDB-29580: 
Structure of HIV-1 BG505 SOSIP-HT2 in complex with two CD4 molecules (class I)
Method: single particle / : Fan C, Dam KA, Bjorkman PJ

PDB-8fyi: 
Structure of HIV-1 BG505 SOSIP-HT1 in complex with one CD4 molecule
Method: single particle / : Fan C, Dam KA, Bjorkman PJ

PDB-8fyj: 
Structure of HIV-1 BG505 SOSIP-HT2 in complex with two CD4 molecules (class I)
Method: single particle / : Fan C, Dam KA, Bjorkman PJ

EMDB-34806: 
SARS-CoV-2 Delta Spike in complex with FP-12A
Method: single particle / : Chen X, Wu YM

EMDB-34807: 
SARS-CoV-2 Delta Spike in complex with IS-9A
Method: single particle / : Mohapatra A, Wu YM

EMDB-34808: 
SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A
Method: single particle / : Chen X, Mohapatra A, Wu YM

PDB-8hhy: 
SARS-CoV-2 Delta Spike in complex with IS-9A
Method: single particle / : Mohapatra A, Wu YM

PDB-8hhz: 
SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A
Method: single particle / : Chen X, Mohapatra A, Wu YM

EMDB-26878: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-3
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26879: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-6
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26880: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-28
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26881: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-31
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26882: 
Structure of the SARS-CoV-2 Omicron BA.1 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-31
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26883: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-34
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26884: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, HSW-1
Method: single particle / : Fan C, Bjorkman PJ

EMDB-26885: 
Structure of the SARS-CoV-2 S S1 doamin in complex with the mouse antibody Fab fragment, HSW-2
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz4: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-3
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz5: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-6
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz6: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-28
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz7: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-31
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz8: 
Structure of the SARS-CoV-2 Omicron BA.1 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-31
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uz9: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, M8a-34
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uza: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the mouse antibody Fab fragment, HSW-1
Method: single particle / : Fan C, Bjorkman PJ

PDB-7uzb: 
Structure of the SARS-CoV-2 S S1 doamin in complex with the mouse antibody Fab fragment, HSW-2
Method: single particle / : Fan C, Bjorkman PJ

EMDB-27208: 
anti-HIV-1 gp120-sCD4 complex antibody CG10 Fab in complex with B41-sCD4
Method: single particle / : Yang Z, Bjorkman PJ, Gershoni JM

EMDB-27209: 
Trimeric HIV-1 Env BG505 SOSIP.664 in complex with sCD4 and antibody 1393A
Method: single particle / : Yang Z, Bjorkman PJ

EMDB-27210: 
HIV-1 Env trimer BG505 SOSIP.664 trimer in complex with sCD4 and 1361 Fab
Method: single particle / : Yang Z, Bjorkman PJ

EMDB-27211: 
HIV-1 Env trimer BG505 SOSIP in complex with sCD4 and 697D Fab
Method: single particle / : Yang Z, Bjorkman PJ

EMDB-27212: 
HIV-1 Env trimer BG505 SOSIP in complex with sCD4 and antibody 830A
Method: single particle / : Yang Z, Bjorkman PJ

PDB-8d5c: 
anti-HIV-1 gp120-sCD4 complex antibody CG10 Fab in complex with B41-sCD4
Method: single particle / : Yang Z, Bjorkman PJ, Gershoni JM

EMDB-26491: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 2
Method: single particle / : Dam KA, Bjorkman PJ

EMDB-26494: 
Cryo-EM map of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 2
Method: single particle / : Dam KA, Bjorkman PJ

EMDB-26495: 
Cryo-EM map of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 3
Method: single particle / : Dam KA, Bjorkman PJ
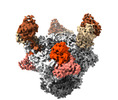
EMDB-26493: 
Cryo-EM structure of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 1
Method: single particle / : Dam KA, Bjorkman PJ

EMDB-26496: 
Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664
Method: single particle / : Dam KA, Bjorkman PJ

PDB-7ugp: 
Cryo-EM structure of BG24 Fabs with an inferred germline light chain and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 containing the N276 gp120 glycan- Class 1
Method: single particle / : Dam KA, Bjorkman PJ

PDB-7ugq: 
Cryo-EM structure of BG24 Fabs with an inferred germline CDRL1 and 10-1074 Fabs in complex with HIV-1 Env 6405-SOSIP.664
Method: single particle / : Dam KA, Bjorkman PJ
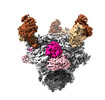
EMDB-26490: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 1
Method: single particle / : Dam KA, Bjorkman PJ
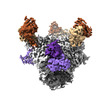
EMDB-26492: 
Cryo-EM structure of BG24 inferred germline Fabs with mature CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1
Method: single particle / : Dam KA, Bjorkman PJ

PDB-7ugn: 
Cryo-EM structure of BG24 inferred germline Fabs with germline CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1 - Class 1
Method: single particle / : Dam KA, Bjorkman PJ

PDB-7ugo: 
Cryo-EM structure of BG24 inferred germline Fabs with mature CDR3s and 10-1074 Fabs in complex with HIV-1 Env immunogen BG505-SOSIPv4.1-GT1
Method: single particle / : Dam KA, Bjorkman PJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
