-Search query
-Search result
Showing all 44 items for (author: berman & h)

EMDB-27500: 
EBNA1 DNA binding domain (DBD) (458-617)+2 repeats of family repeat (FR) region
Method: single particle / : Mei Y, Lieberman PM, Murakami K

PDB-8dlf: 
EBNA1 DNA binding domain (DBD) (458-617)+2 repeats of family repeat (FR) region
Method: single particle / : Mei Y, Lieberman PM, Murakami K
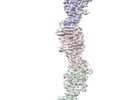
EMDB-23481: 
Cryo-EM structure of OmcZ nanowire from Geobacter sulfurreducens
Method: helical / : Gu Y, Srikanth V, Malvankar NS, Samatey FA

PDB-7lq5: 
Cryo-EM structure of OmcZ nanowire from Geobacter sulfurreducens
Method: helical / : Gu Y, Srikanth V, Malvankar NS, Samatey FA

EMDB-13453: 
Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

EMDB-13454: 
Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

PDB-7piu: 
Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

PDB-7piv: 
Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution.
Method: single particle / : Heyder NA, Schmidt A, Kleinau G, Hilal T, Scheerer P

EMDB-24077: 
CryoEM structure of neutralizing nanobody Nb30 in complex with SARS-CoV2 spike
Method: single particle / : Xu K, Kwong PD

EMDB-24078: 
CryoEM structure of neutralizing nanobody Nb12 in complex with SARS-CoV2 spike
Method: single particle / : Xu K, Kwong PD

PDB-7my2: 
CryoEM structure of neutralizing nanobody Nb30 in complex with SARS-CoV2 spike
Method: single particle / : Xu K, Kwong PD

PDB-7my3: 
CryoEM structure of neutralizing nanobody Nb12 in complex with SARS-CoV2 spike
Method: single particle / : Xu K, Kwong PD

EMDB-21161: 
Pore-forming protein
Method: single particle / : Xia S, Ruan J, Wu H
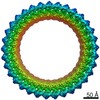
EMDB-21160: 
Gasdermin D pore
Method: single particle / : Xia S, Ruan J, Wu H

EMDB-21514: 
map of the mammalian Mediator complex
Method: single particle / : Zhao H, Young N, Asturias F

PDB-6w1s: 
Atomic model of the mammalian Mediator complex
Method: single particle / : Zhao H, Young N, Asturias F

EMDB-23393: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C601
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-23394: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C603
Method: single particle / : Yang Z, Barnes CO, Bjorkman PJ

EMDB-23395: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C643
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-23396: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C663
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-23397: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C666
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-23398: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C669
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-23399: 
Structure of the SARS-CoV-2 spike trimer in complex with mRNA vaccine induced neutralizing antibody C670
Method: single particle / : Yang Z, Barnes CO, Bjorkman PJ

EMDB-10417: 
GroEL map obtained using the Preassis method for grid preparation
Method: single particle / : Zhao J, Xu H, Carroni M, Zou X

EMDB-10466: 
Cryo-ET on GroEL
Method: electron tomography / : Zou XD, Zhao JJ, Xu HY, Carroni M

EMDB-22681: 
SARS-COV-2 nsp1 in complex with human 40S ribosome
Method: single particle / : Wang L, Shi M, Wu H

PDB-7k5i: 
SARS-COV-2 nsp1 in complex with human 40S ribosome
Method: single particle / : Wang L, Shi M, Wu H

EMDB-10462: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408
Method: single particle / : Srinivas H

EMDB-10463: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib
Method: single particle / : Srinivas H

PDB-6tcz: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408
Method: single particle / : Srinivas H

PDB-6td5: 
Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib
Method: single particle / : Srinivas H

EMDB-20391: 
CryoEM structure of mouse mediator subunit one deletion
Method: single particle / : Asturias F, Zhao H, Young N

EMDB-20392: 
CryoEM structure of wild type mouse mediator with MED19-FLAG
Method: single particle / : Asturias F, Zhao H

EMDB-20393: 
CryoEM structure of wild type mediator with MED25-FLAG
Method: single particle / : Asturias F, Zhao H

EMDB-7450: 
Cryo-EM structure of the Gasdermin A3 membrane pore
Method: single particle / : Ruan J, Xia S, Wu H

EMDB-7451: 
Cryo-EM structure of the Gasdermin A3 membrane pore
Method: single particle / : Ruan J, Xia S, Wu H

EMDB-7449: 
Cryo-EM structure of the Gasdermin A3 membrane pore
Method: single particle / : Ruan J, Wu H
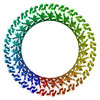
PDB-6cb8: 
Cryo-EM structure of the Gasdermin A3 membrane pore
Method: single particle / : Ruan J, Wu H

EMDB-2103: 
Heritable yeast prions have a highly organized 3-dimensional architecture with inter-fiber structures
Method: subtomogram averaging / : Frangakis A, Saibil HR

EMDB-2104: 
Heritable yeast prions have a highly organized 3-dimensional architecture with inter-fiber structures
Method: subtomogram averaging / : Frangakis A, Saibil HR

PDB-3iyd: 
Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Method: single particle / : Hudson BP, Quispe J, Lara S, Kim Y, Berman H, Arnold E, Ebright RH, Lawson CL

EMDB-5127: 
Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Method: single particle / : Hudson BP, Quispe J, Lara S, Kim Y, Berman HM, Arnold E, Ebright RH, Lawson CL

EMDB-1050: 
The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a two-fold symmetric, 14-subunit spiral.
Method: single particle / : Sewell BT, Berman MN, Meyers PR, Jandhyala D, Benedik MJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
