-Search query
-Search result
Showing all 41 items for (author: bator & cm)

EMDB-41037: 
Cryo-EM structure of immature Zika virus
Method: single particle / : Moustafa IM, Hafenstein SL
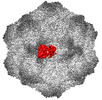
EMDB-26786: 
CPV Affinity Purified Polyclonal Fab A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
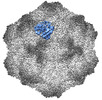
EMDB-26787: 
CPV Affinity Purified Polyclonal Fab B Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
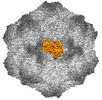
EMDB-26788: 
CPV Total-Fab Polyclonal A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
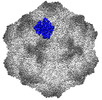
EMDB-26789: 
CPV Total-Fab Polyclonal B Site Fab (1 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
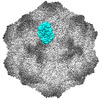
EMDB-26790: 
CPV Total-Fab Polyclonal B Site Fab (2 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

EMDB-27943: 
9H2 Fab-poliovirus 1 complex
Method: single particle / : Charnesky AJ
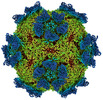
EMDB-27947: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ

EMDB-27948: 
9H2 Fab-poliovirus 2 complex
Method: single particle / : Charnesky AJ
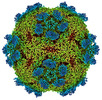
EMDB-27949: 
9H2 Fab-Sabin poliovirus 3 complex
Method: single particle / : Charnesky AJ
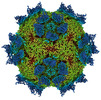
EMDB-27950: 
9H2 Fab-Sabin poliovirus 2 complex
Method: single particle / : Charnesky AJ
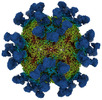
EMDB-27951: 
9H2 Fab-Sabin poliovirus 1 complex
Method: single particle / : Charnesky AJ

EMDB-27397: 
Gokushovirus EC6098
Method: single particle / : Lee H, Fane BA, Hafenstein SL

EMDB-24741: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

EMDB-23656: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23657: 
Canine parvovirus and Fab14 at partial occupancy
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23658: 
Canine parvovirus and Fab14 asymmetric reconstruction
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23659: 
Canine parvovirus asymmetric map
Method: single particle / : Goteschius DJ, Hartmann SR, Hafenstein SL

EMDB-23081: 
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility
Method: single particle / : Hartmann SR, Goetschius DJ

EMDB-22645: 
Murine polyomavirus (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22646: 
Murine polyomavirus with 8A7H5 Fab (icosahedral reconstruction)
Method: single particle / : Goetschius DJ, Hafenstein SL

EMDB-22526: 
Subparticle Map of ZIKV MR-766
Method: single particle / : Jose J, Hafenstein S

EMDB-22527: 
Icosahedral Map of ZIKV MR-766
Method: single particle / : Jose J, Hafenstein S

EMDB-20766: 
Structure of the EVA71 strain 11316 capsid
Method: single particle / : Lee H, Hafenstein S

EMDB-20768: 
EV-A71 strain 11316 complexed with MADAL compound 22
Method: single particle / : Lee H, Hafenstein S

EMDB-20769: 
EV-A71 strain 11316 complexed with MADAL compound 30
Method: single particle / : Lee H, Hafenstein S

EMDB-20624: 
Empty AAV3B
Method: single particle / : Subramanian S, Hafenstein S

EMDB-20625: 
AAV3B genome containing
Method: single particle / : Subramanian S, Hafenstein S

EMDB-20001: 
Structure of canine parvovirus in complex with transferrin receptor type-1
Method: single particle / : Lee H, Hafenstein S

EMDB-20002: 
Structure of canine parvovirus in complex with transferrin receptor type-1
Method: single particle / : Lee H, Hafenstein S

EMDB-20003: 
Transferrin-transferrin receptor complex bound to canine parvovirus capsid
Method: single particle / : Lee H, Hafenstein S

EMDB-9363: 
Structure of M. spretus Endogenous Virus Element (EVE) Virus-like particle (VLP)
Method: single particle / : Callaway HM, Subramanian S

EMDB-5179: 
The Interaction of Decay-accelerating Factor with Echovirus 7
Method: single particle / : PLEVKA P, HAFENSTEIN S, ZHANG Y, HARRIS KG, CIEFUENTE JO, BOWMAN VD, CHIPMAN PR, LIN F, MEDOF DE, BATOR CM, ROSSMANN MG

EMDB-1562: 
CryoEM reconstructions of PV3 complexed with deglycosylated CD155
Method: single particle / : Zhang P, Mueller S, Morais MC, Bator-Kelly CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG

EMDB-1563: 
CryoEM reconstructions of PV2 complexed with deglycosylated CD155
Method: single particle / : Zhang P, Mueller S, Morais MC, Bator-Kelly CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG

EMDB-1570: 
CryoEM reconstructions of PV1 complexed with deglycosylated CD155
Method: single particle / : Zhang P, Mueller S, Morais MC, Bator-Kelly CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG

EMDB-1411: 
Interaction of decay-accelerating factor with coxsackievirus B3.
Method: single particle / : Hafenstein S

EMDB-1412: 
Interaction of decay-accelerating factor with coxsackievirus B3.
Method: single particle / : Hafenstein S

EMDB-1166: 
Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN.
Method: single particle / : Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG
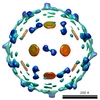
EMDB-1167: 
Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN.
Method: single particle / : Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG

EMDB-1114: 
The crystal structure of coxsackievirus A21 and its interaction with ICAM-1.
Method: single particle / : Xiao C, Bator-Kelly CM, Rieder E, Chipman PR, Craig A, Kuhn RJ, Wimmer E, Rossmann MG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
