-Search query
-Search result
Showing all 35 items for (author: asano & s)
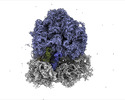
EMDB-35411: 
Dibekacin-bound E.coli 70S ribosome in the PURE system
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T
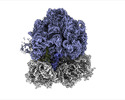
EMDB-35412: 
Arbekacin-bound E.coli 70S ribosome in the PURE system
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T
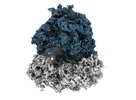
EMDB-35413: 
Dibekacin-added human 80S ribosome
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T
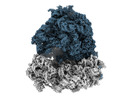
EMDB-35414: 
Arbekacin-added human 80S ribosome
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T

PDB-8ifb: 
Dibekacin-bound E.coli 70S ribosome in the PURE system
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T

PDB-8ifc: 
Arbekacin-bound E.coli 70S ribosome in the PURE system
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T

PDB-8ifd: 
Dibekacin-added human 80S ribosome
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T

PDB-8ife: 
Arbekacin-added human 80S ribosome
Method: single particle / : Tomono J, Asano K, Chiashi T, Tanaka Y, Yokoyama T
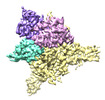
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
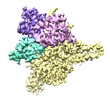
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

PDB-8e4y: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H

PDB-8e50: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

EMDB-14313: 
The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

EMDB-14314: 
The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

EMDB-14315: 
The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

PDB-7r4i: 
The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

PDB-7r4q: 
The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

PDB-7r4r: 
The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody
Method: single particle / : Casasnovas JM, Melero R, Arranz R, Fernandez LA

EMDB-22283: 
cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970
Method: single particle / : Sun B, Kobilka BK, Sloop KW, Feng D, Kobilka TS

PDB-6xox: 
cryo-EM of human GLP-1R bound to non-peptide agonist LY3502970
Method: single particle / : Sun B, Kobilka BK, Sloop KW, Feng D, Kobilka TS

EMDB-4392: 
In situ subtomogram average of a bacterial encapsulin expressed in HEK293T cells (no iron treatment)
Method: subtomogram averaging / : Erdmann PS, Plitzko JM

EMDB-4393: 
In situ subtomogram average of a bacterial encapsulin expressed in HEK293T cells (after iron treatment)
Method: subtomogram averaging / : Erdmann PS, Plitzko JM

EMDB-3967: 
In situ cryo-electron tomogram from Chlamydomonas reinhardtii of the cellular environment around the nuclear envelope
Method: electron tomography / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3932: 
In situ subtomogram average of the Chlamydomonas double-capped 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3933: 
In situ subtomogram average of the Chlamydomonas single-capped 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3934: 
In situ subtomogram average of the Chlamydomonas ground state 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3935: 
In situ subtomogram average of the Chlamydomonas processing state 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3936: 
In situ subtomogram average of the Chlamydomonas membrane-tethered 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3937: 
In situ subtomogram average of the Chlamydomonas basket-tethered 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3938: 
In situ subtomogram average of the Chlamydomonas free 26S proteasome
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3939: 
Local refinement of the extra density that tethers 26S proteasomes to the NPC basket
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3940: 
Local refinement of the extra density that tethers 26S proteasomes to the inner nuclear membrane
Method: subtomogram averaging / : Albert S, Schaffer M, Beck F, Mosalaganti S, Asano S, Thomas HF, Plitzko J, Beck M, Baumeister W, Engel BD

EMDB-3100: 
Cryo-electron tomography of a Golgi intracisternal protein array
Method: subtomogram averaging / : Engel BD, Schaffer M, Albert S, Asano S, Plitzko JM, Baumeister W

EMDB-2830: 
Electron cryotomography, subtomogram averaging and classification of 26S proteasomes in situ in intact hippocampal neurons
Method: subtomogram averaging / : Asano S, Fukuda Y, Beck F, Aufderheide A, Foerster F, Danev R, Baumeister W

EMDB-2831: 
Electron cryotomography, subtomogram averaging and classification of 26S proteasomes in situ in intact hippocampal neurons
Method: subtomogram averaging / : Asano S, Fukuda Y, Beck F, Aufderheide A, Foerster F, Danev R, Baumeister W
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
