-Search query
-Search result
Showing all 33 items for (author: arias-palomo & e)

EMDB-16521: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

EMDB-16524: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

EMDB-16531: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E, Araujo-Bazan L, Berger JM

PDB-8ca9: 
Cryo-EM structure of the Cibeles-Demetra 3:3 heterocomplex from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E

PDB-8cad: 
Cryo-EM structure of the Ceres homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E

PDB-8can: 
Cryo-EM structure of the Cora homohexamer from Galleria mellonella saliva
Method: single particle / : Spinola-Amilibia M, Arias-Palomo E

EMDB-15848: 
IstA transposase cleaved donor complex
Method: single particle / : Spinola-Amilibia M, de la Gandara A, Araujo-Bazan L, Berger JM, Arias-Palomo E

PDB-8b4h: 
IstA transposase cleaved donor complex
Method: single particle / : Spinola-Amilibia M, de la Gandara A, Araujo-Bazan L, Berger JM, Arias-Palomo E

EMDB-10492: 
Vip3Aa protoxin structure
Method: single particle / : Nunez-Ramirez R, Huesa J, Bel Y, Ferre J, Casino P, Arias-Palomo E

EMDB-10493: 
Vip3Aa toxin structure
Method: single particle / : Nunez-Ramirez R, Huesa J, Bel Y, Ferre J, Casino P, Arias-Palomo E

PDB-6tfj: 
Vip3Aa protoxin structure
Method: single particle / : Nunez-Ramirez R, Huesa J, Bel Y, Ferre J, Casino P, Arias-Palomo E

PDB-6tfk: 
Vip3Aa toxin structure
Method: single particle / : Nunez-Ramirez R, Huesa J, Bel Y, Ferre J, Casino P, Arias-Palomo E

EMDB-4537: 
E. coli DnaBC apo complex
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM

EMDB-4538: 
E. coli DnaBC complex bound to ssDNA
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM

PDB-6qel: 
E. coli DnaBC apo complex
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM

PDB-6qem: 
E. coli DnaBC complex bound to ssDNA
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM

EMDB-3031: 
Cryo-EM structure of ATP-bound IstB in complex to duplex DNA
Method: single particle / : Arias-Palomo E, Berger JM
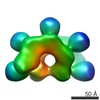
EMDB-3032: 
Cryo-EM structure of ATP-bound IstB
Method: single particle / : Arias-Palomo E, Berger JM

EMDB-2508: 
Nucleotide and partner-protein control of bacterial replicative helicase structure and function
Method: single particle / : Strycharska MS, Arias-Palomo E, Lyubimov AY, Erzberger JP, O'Shea VL, Bustamante C, Berger JM

EMDB-2198: 
Architecture of human translation initiation factor 3
Method: single particle / : Querol-Audi J, Sun C, Mortimer S, Arias-Palomo E, Vogan J, Doudna J, Nogales E, Cate J
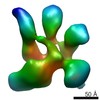
EMDB-2199: 
Architecture of human translation initiation factor 3
Method: single particle / : Querol-Audi J, Sun C, Mortimer S, Arias-Palomo E, Vogan J, Doudna J, Nogales E, Cate J

EMDB-2321: 
The bacterial DnaC helicase loader is a DnaB ring breaker
Method: single particle / : Arias-Palomo E, O'Shea VL, Hood IV, Berger JM

EMDB-2322: 
The bacterial DnaC helicase loader is a DnaB ring breaker
Method: single particle / : Arias-Palomo E, O'Shea VL, Hood IV, Berger JM

EMDB-1851: 
EM map of the negative stained SMG-1-9 complex.
Method: single particle / : Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O

EMDB-1852: 
EM map of the negative stained SMG-1-8-9 complex.
Method: single particle / : Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O

EMDB-1853: 
Cryo-EM map of the SMG-1-9 complex.
Method: single particle / : Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O

EMDB-1360: 
Structure of TOR and its complex with KOG1.
Method: single particle / : Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O

EMDB-1361: 
Structure of TOR and its complex with KOG1.
Method: single particle / : Adami A, Garcia-Alvarez B, Arias-Palomo E, Barford D, Llorca O

EMDB-2865: 
Architecture of the yeast pontin-reptin complex
Method: single particle / : Torreira E, Jha S, Lopez-Blanco JR, Arias-Palomo E, Chacon P, Canas C, Ayora S, Dutta A, Llorca O

EMDB-1510: 
Structure of full-length Epac2 in complex with cyclic-AMP and Rap.
Method: single particle / : Arias-Palomo E, Rehmann H, Hadders M, Schwede F, Bos JL, Llorca O

EMDB-1103: 
Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3.
Method: single particle / : Llorca O, Arias-Palomo E, Zugaza JL, Bustelo XR

EMDB-1104: 
Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3.
Method: single particle / : Llorca O, Arias-Palomo E, Zugaza JL, Bustelo XR
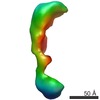
EMDB-1105: 
Global conformational rearrangements during the activation of the GDP/GTP exchange factor Vav3.
Method: single particle / : Llorca O, Arias-Palomo E, Zugaza JL, Bustelo XR
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
