-Search query
-Search result
Showing 1 - 50 of 72 items for (author: anne & s & meyer)

EMDB-15774: 
p97 with I3 substrate in channel
Method: single particle / : Saibil HR, van den Boom J, Marini G, Meyer H

EMDB-15778: 
p97-substrate complex with SPI substrate on subunit D
Method: single particle / : Saibil HR, van den Boom J, Marini G, Meyer H

EMDB-15846: 
p97-SPI substrate complex on subunit E
Method: single particle / : Saibil H, van den Boom J, Marini G, Meyer H

EMDB-15847: 
p97 complexed with SPI substrate on subunit C
Method: single particle / : Saibil HR, van den Boom J, Marini G, Meyer H

EMDB-15861: 
p97-p37-SPI substrate complex
Method: single particle / : van den Boom J, Marini G, Meyer H, Saibil H

PDB-8b5r: 
p97-p37-SPI substrate complex
Method: single particle / : van den Boom J, Marini G, Meyer H, Saibil H

EMDB-27973: 
Erwinia amylovora 70S Ribosome
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-27993: 
Erwinia amlyavora 50S ribosome
Method: subtomogram averaging / : Prichard A, Lee J, Laughlin TG, Lee A, Thomas K, Sy A, Spencer T, Asavavimol A, Cafferata A, Cameron M, Chiu N, Davydov D, Desai I, Diaz G, Guereca M, Hearst K, Huang L, Jacobs E, Johnson A, Kahn S, Koch R, Martinez A, McDonald-Uyeno A, Norquist M, Pau T, Prasad G, Saam K, Sandu M, Sarabia A, Schumaker S, Sonin A, Zhao A, Corbett A, Pogliano K, Meyer J, Grose J, Villa E, Dutton R, Pogliano J

EMDB-28003: 
Erwinia phage vB_EamM_RAY (RAY) Capsid Vertex
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28004: 
Erwinia phage vB_EamM_RAY (RAY) Capsid Collar
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28005: 
Erwinia phage vB_EamM_RAY (RAY) Tail Sheath
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28006: 
Erwinia phage vB_EamM_RAY (RAY) Baseplate
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28007: 
Erwinia phage vB_EamM_RAY (RAY) Chimallin
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28008: 
Erwinia phage vB_EamM_RAY (RAY) Putative PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28009: 
Pseudomonas chlororaphis phage 201phi2-1 PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-28010: 
Pseudomonas phage phiKZ PhuZ Filament
Method: subtomogram averaging / : Laughlin TG, Villa E

EMDB-23542: 
Structure of full-length GluK1 with L-Glu
Method: single particle / : Meyerson JR, Selvakumar P

PDB-7lvt: 
Structure of full-length GluK1 with L-Glu
Method: single particle / : Meyerson JR, Selvakumar P

PDB-7l6o: 
Cryo-EM structure of HIV-1 Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23518: 
Cryo-EM map of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lu9: 
Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23519: 
Cryo-EM map of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Edwards RJ, Manne K, Acharya P

PDB-7lua: 
Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23152: 
Cryo-electron microscopy reconstruction of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer
Method: single particle / : Edwards RJ, Acharya P

EMDB-23153: 
Cryo-electron microscopy local refinement of antibody DH898.1 Fab-dimer bound to glycans 332, 392, and 396 of HIV Env CH848 10.17 SOSIP trimer
Method: single particle / : Edwards RJ, Acharya P

EMDB-23149: 
Cryo-electron microscopy reconstruction of antibody DH898.1 Fab-dimer bound near the CD4 binding site of HIV Env SOSIP trimer CH848 10.17
Method: single particle / : Edwards RJ, Acharya P

EMDB-12187: 
SARS-CoV-2 RBD-62 in complex with ACE2 peptidase domain
Method: single particle / : Elad N, Dym O, Zahradnik J, Schreiber G

PDB-7bh9: 
SARS-CoV-2 RBD-62 in complex with ACE2 peptidase domain
Method: single particle / : Elad N, Dym O, Zahradnik J, Schreiber G

EMDB-23124: 
Cryo-electron microcospy reconstruction of CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 HIV Env
Method: single particle / : Edwards RJ, Acharya P

EMDB-23145: 
Cryo-electron microscopy reconstruction of locally refined antibody DH898.1 Fab-dimer
Method: single particle / : Edwards RJ, Acharya P

PDB-7l6m: 
Cryo-EM structure of DH898.1 Fab-dimer from local refinement of the Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer
Method: single particle / : Manne K, Edwards RJ, Acharya P

EMDB-23094: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-23095: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-23097: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l02: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to one copy of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l06: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound to two copies of domain-swapped antibody 2G12
Method: single particle / : Manne K, Henderson R, Acharya P

PDB-7l09: 
Cryo-EM structure of SARS-CoV-2 2P S ectodomain bound domain-swapped antibody 2G12 from masked 3D refinement
Method: single particle / : Manne K, Henderson R, Acharya P

EMDB-10432: 
In situ ER membrane bound ribosome
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10433: 
In situ ER membrane bound translocon-ribosome
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10434: 
In situ ER membrane bound ribosome-free translocon
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10435: 
Putative in situ ER membrane bound major histocompatibility peptide loading complex (PLC)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S
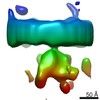
EMDB-10436: 
Putative in situ ER membrane bound major histocompatibility peptide loading complex (PLC)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S
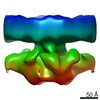
EMDB-10437: 
Putative in situ ER membrane bound inositol trisphosphate receptor complex (IP3R)
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10439: 
Example of tomogram used for in situ template-free membrane bound complexes determination
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Chakraborty S

EMDB-10449: 
ER Microsome in Fig. 3 (Martinez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-10450: 
Endoplasmic Reticulum microsome used for Videos in (Martinez-Sanchez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-10451: 
ER microsome released with the code in (Martinez-Sanchez et al., Nature Methods)
Method: electron tomography / : Martinez-Sanchez A, Lucic V, Pfeffer S, Forster F

EMDB-0074: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0075: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V

EMDB-0084: 
Template-free detection and classification of microsomal membrane bound complexes
Method: subtomogram averaging / : Martinez-Sanchez A, Lucic V
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
