-Search query
-Search result
Showing 1 - 50 of 185 items for (author: andrew & p & carter)

EMDB-17835: 
Consensus cryo-EM structure of Dynein-Dynactin-JIP3(1-185)-LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17836: 
Consensus cryo-EM structure of Dynein-dynactin-JIP3(1-560)-LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17825: 
Cytoplasmic dynein-1 motor domain in post-powerstroke state
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17826: 
Cytoplasmic dynein-1 motor domain bound to dynactin-p150glued and LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17828: 
Cytoplasmic dynein-1 motor domain bound to LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17829: 
Cytoplasmic dynein-1 A1/A2 motor domains bound to LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17830: 
Cytoplasmic dynein-A heavy chain bound to dynactin-p150glued and IC-LC tower
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17831: 
Cytoplasmic dynein-B heavy chain bound to IC-LC tower
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17832: 
Cytoplasmic dynein-1 heavy chain bound to JIP3-LZI
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17833: 
Cytoplasmic dynein-1 heavy chain bound to JIP3-RH1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17834: 
Dynactin pointed end bound to JIP3
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-17873: 
Composite structure of Dynein-Dynactin-JIP3-LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pqv: 
Cytoplasmic dynein-1 motor domain in post-powerstroke state
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pqw: 
Cytoplasmic dynein-1 motor domain bound to dynactin-p150glued and LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pqy: 
Cytoplasmic dynein-1 motor domain bound to LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pqz: 
Cytoplasmic dynein-1 A1/A2 motor domains bound to LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr0: 
Cytoplasmic dynein-A heavy chain bound to dynactin-p150glued and IC-LC tower
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr1: 
Cytoplasmic dynein-B heavy chain bound to IC-LC tower
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr2: 
Cytoplasmic dynein-1 heavy chain bound to JIP3-LZI
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr3: 
Cytoplasmic dynein-1 heavy chain bound to JIP3-RH1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr4: 
Dynactin pointed end bound to JIP3
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8pr5: 
Structure of the autoinhibited dynactin p150glued projection
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

PDB-8ptk: 
Composite structure of Dynein-Dynactin-JIP3-LIS1
Method: single particle / : Singh K, Lau CK, Manigrasso G, Gassmann R, Carter AP

EMDB-18475: 
Cryo-electron tomogram of an induced S2 cell protrusion. The cell was treated with 2uM thapsigargin (5h) and with 2.5uM Cytochalasin D (2h).
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-29680: 
C6HR1_8r: Extendable repeat protein heptamer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29847: 
C3HR3_8r: Extendable repeat protein trimer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29849: 
C6HR1_4r: Extendable repeat protein hexamer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29851: 
C4HR1_8r_shift5: Extendable repeat protein fiber
Method: helical / : Bethel NP, Borst AJ

EMDB-29856: 
C3HR3_9r_shift4: Extendable repeat protein fiber
Method: helical / : Bethel NP, Borst AJ

EMDB-29894: 
C4HR1_4r: Extendable repeat protein tetramer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29904: 
C5HR2_4r: Extendable repeat protein pentamer
Method: single particle / : Bethel NP, Borst AJ

PDB-8ga9: 
C4HR1_4r: Extendable repeat protein tetramer
Method: single particle / : Bethel NP, Borst AJ
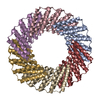
PDB-8gaa: 
C6HR1_4r: Extendable repeat protein hexamer
Method: single particle / : Bethel NP, Borst AJ, Baker D

PDB-8gaq: 
C5HR2_4r: Extendable repeat protein pentamer
Method: single particle / : Bethel NP, Borst AJ

EMDB-16877: 
Subtomogram averaging structure of cofilactin filament inside microtubule lumen of Drosophila S2 cell protrusion.
Method: subtomogram averaging / : Ventura Santos C, Carter AP

PDB-8oh4: 
Subtomogram averaging structure of cofilactin filament inside microtubule lumen of Drosophila S2 cell protrusion.
Method: subtomogram averaging / : Ventura Santos C, Carter AP

EMDB-16685: 
Tomogram of an induced protrusion of a Drosophila S2 cell
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-16693: 
Tomogram of an induced protrusion of a Drosophila S2 cell
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-16695: 
Tomogram of an induced protrusion of a Drosophila S2 alpha-tubulin acetyltransferase knock-out (dTAT KO) cell with a filament inside the microtubule lumen
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-16720: 
Tomogram of an induced protrusion of a Drosophila S2 cell with filaments inside the microtubule lumen.
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-16800: 
Tomogram of an induced protrusion of a cofilin knock-down Drosophila S2 cell with filaments inside the microtubule lumen
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-16811: 
Tomogram of an induced protrusion of a cofilin knock-down Drosophila S2 cell with a filament inside the microtubule lumen.
Method: electron tomography / : Ventura Santos C, Carter AP

EMDB-28862: 
KWOCA 4 nanoparticle
Method: single particle / : Antanasijevic A, Ward AB

EMDB-28929: 
Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains - KWOCA51
Method: single particle / : Borst AJ, King NP

EMDB-33748: 
Spike_GSAS_6P protomer RBD domain bound with R1-32 Fab and ACE2 with 3:3:3 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
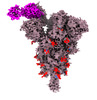
EMDB-33760: 
Spike_GSAS_6P and R1-32 Fab with 3to1 ratio
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
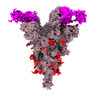
EMDB-33764: 
SARS-COV-2 Spike_GSAS_6P bound with two R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
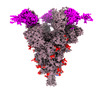
EMDB-33766: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X

EMDB-33772: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2
Method: single particle / : Liu B, Gao X, Li Z, Chen X, He J, Chen L, Xiong X
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
