-Search query
-Search result
Showing 1 - 50 of 166 items for (author: alexandre & t)
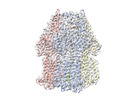
EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution
Method: single particle / : Boernsen C, Mueller RT, Pos KM, Frangakis AS
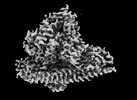
EMDB-41617: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M, Sun M

PDB-8tu6: 
CryoEM structure of PI3Kalpha
Method: single particle / : Valverde R, Shi H, Holliday M
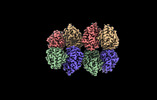
EMDB-28013: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)
Method: single particle / : Ambrosio AL, Dias SM, Quesnay JE, Portugal RV, Cassago A, van Heel MG, Islam Z, Rodrigues CT

PDB-8ec6: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)
Method: single particle / : Ambrosio AL, Dias SM, Quesnay JE, Portugal RV, Cassago A, van Heel MG, Islam Z, Rodrigues CT

EMDB-15113: 
Human mature large subunit of the ribosome with eIF6 and homoharringtonine bound
Method: single particle / : Faille A, Warren AJ, Dent KC

PDB-8a3d: 
Human mature large subunit of the ribosome with eIF6 and homoharringtonine bound
Method: single particle / : Faille A, Warren AJ, Dent KC

EMDB-27971: 
V-K CAST Transpososome from Scytonema hofmanni, major configuration
Method: single particle / : Rizo AN, Park JU, Tsai AW, Kellogg EH
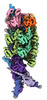
EMDB-27972: 
V-K CAST Transpososome from Scytonema hofmanni, minor configuration
Method: single particle / : Rizo AR, Park JU, Tsai AW, Kellogg EK

PDB-8ea3: 
V-K CAST Transpososome from Scytonema hofmanni, major configuration
Method: single particle / : Rizo AN, Park JU, Tsai AW, Kellogg EH

PDB-8ea4: 
V-K CAST Transpososome from Scytonema hofmanni, minor configuration
Method: single particle / : Rizo AR, Park JU, Tsai AW, Kellogg EK

EMDB-25453: 
TnsBctd-TnsC-TniQ complex
Method: single particle / : Park J, Tsai AWT, Kellogg EH

PDB-7svu: 
TnsBctd-TnsC-TniQ complex
Method: single particle / : Park J, Tsai AWT, Kellogg EH

EMDB-26936: 
Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab
Method: single particle / : Kucharska I, Prieto K, Rubinstein JL, Julien JP

PDB-7v05: 
Complex of Plasmodium falciparum circumsporozoite protein with 850 Fab
Method: single particle / : Kucharska I, Prieto K, Rubinstein JL, Julien JP

EMDB-26121: 
Skd3_ATPyS_FITC-casein Hexamer, AAA+ only
Method: single particle / : Rizo AN

EMDB-26122: 
Skd3, hexamer, filtered
Method: single particle / : Rizo AN, Cupo RR

EMDB-27730: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432
Method: single particle / : QCRG Structural Biology Consortium, Remesh SG, Merz GE, Brilot AF, Chio U, Verba KA

EMDB-27731: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293
Method: single particle / : QCRG Structural Biology Consortium, Remesh SG, Merz GE, Brilot AF, Chio U, Verba KA

PDB-8dv1: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432
Method: single particle / : QCRG Structural Biology Consortium, Remesh SG, Merz GE, Brilot AF, Chio U, Verba KA

PDB-8dv2: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293
Method: single particle / : QCRG Structural Biology Consortium, Remesh SG, Merz GE, Brilot AF, Chio U, Verba KA

EMDB-13930: 
3D reconstruction of the membrane domains of the sialic acid TRAP transporter HiSiaQM from Haemophilus influenzae in lipid nanodiscs bound to a high affinity megabody
Method: single particle / : Peter MF, Hagelueken G

PDB-7qe5: 
Structure of the membrane domains of the sialic acid TRAP transporter HiSiaQM from Haemophilus influenzae
Method: single particle / : Peter MF, Hagelueken G

EMDB-14153: 
SARS-CoV-2 Spike, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

PDB-7qus: 
SARS-CoV-2 Spike, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14152: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14154: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C1 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-14155: 
SARS-CoV-2 Spike, C1 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

PDB-7qur: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry
Method: single particle / : Naismith JH, Yang Y, Liu JW

EMDB-13976: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound)
Method: single particle / : Kao WC, Hunte C

EMDB-13977: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated)
Method: single particle / : Kao WC, Hunte C

PDB-7qhm: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound)
Method: single particle / : Kao WC, Hunte C

PDB-7qho: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated)
Method: single particle / : Kao WC, Hunte C

EMDB-11804: 
Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-11805: 
Structure of Wild-type Human Potassium Chloride Transporter KCC3 in NaCl (MSP E3D1)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-11610: 
CryoEM structure of a beta3K279T GABA(A)R homomer in complex with megabody MbNbF3c7HopQ
Method: single particle / : Uchanski T, Masiulis S

EMDB-4542: 
CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25
Method: single particle / : Uchanski T, Masiulis S, Fischer B, Kalichuk V, Wohlkoening A, Zoegg T, Remaut H, Vranken W, Aricescu AR, Pardon E, Steyaert J

PDB-6qfa: 
CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25
Method: single particle / : Uchanski T, Masiulis S, Fischer B, Kalichuk V, Wohlkoening A, Zoegg T, Remaut H, Vranken W, Aricescu AR, Pardon E, Steyaert J

EMDB-24391: 
Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 7.5
Method: single particle / : Godoy AS, Song Y, Nakamura AM, Noske GD, Gawriljuk VO, Fernandes RS, Oliva G

EMDB-24392: 
Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0
Method: single particle / : Godoy AS, Song Y, Nakamura AM, Noske GD, Gawriljuk VO, Fernandes RS, Oliva G

PDB-7rb0: 
Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 7.5
Method: single particle / : Godoy AS, Song Y, Nakamura AM, Noske GD, Gawriljuk VO, Fernandes RS, Oliva G

PDB-7rb2: 
Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0
Method: single particle / : Godoy AS, Song Y, Nakamura AM, Noske GD, Gawriljuk VO, Fernandes RS, Oliva G

EMDB-12311: 
Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

PDB-7ngb: 
Structure of Wild-Type Human Potassium Chloride Transporter KCC3 in NaCl (LMNG/CHS)
Method: single particle / : Chi G, Man H, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL

EMDB-11799: 
Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Reference Map)
Method: single particle / : Chi G, Man H, Ebenhoch R, Reggiano G, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Singh NK, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, DiMaio F, Duerr KL, Structural Genomics Consortium (SGC)

EMDB-11800: 
Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl (Subclass)
Method: single particle / : Chi G, Man H, Ebenhoch R, Reggiano G, Pike ACW, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Singh NK, Abrusci P, Arrowsmith CH, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, DiMaio F, Duerr KL, Structural Genomics Consortium (SGC)

EMDB-11801: 
Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map)
Method: single particle / : Ebenhoch R, Chi G, Man H, Wang D, McKinley G, Mukhopadhyay SMM, MacLean EM, Chalk R, Moreau C, Snee M, Bohstedt T, Liko I, Tehan BG, Almeida FG, Elkins J, Singh NK, Abrusci P, Arrowsmith CH, Tang H, Robinson CV, Bountra C, Edwards AM, Marsden BD, Burgess-Brown NA, Duerr KL, Structural Genomics Consortium (SGC)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
