-Search query
-Search result
Showing 1 - 50 of 51 items for (author: albert & cupo)

EMDB-22170: 
BG505 SOSIPv5.2 in complex with PGT122 and two RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22171: 
BG505 SOSIPv5.2 in complex with PGT122 and three RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22172: 
C3 symmetric BG505 SOSIPv5.2 in complex with PGT122 and three RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22173: 
BG505 SOSIPv5.2 in complex with PGT122 and one RM20E1 Fab
Method: single particle / : Cottrell CA, Ward AB

EMDB-22178: 
BG505 SOSIPv5.2 in complex with PGT122 and no RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-22179: 
C3 symmetric BG505 SOSIPv5.2 in complex with PGT122 and no RM20E1 Fabs
Method: single particle / : Cottrell CA, Ward AB

EMDB-8714: 
Cryo-EM reconstruction of B41 SOSIP.664
Method: single particle / : Pallesen J, Ozorowski G, de Val N, Ward AB

EMDB-8715: 
Cryo-EM reconstruction of B41 SOSIP.664 in complex with soluble CD4 (D1-D2)
Method: single particle / : Pallesen J, Ozorowski G, de Val N, Ward AB

EMDB-8716: 
Cryo-EM reconstruction of B41 SOSIP.664 in complex with fragment antigen binding of PGV04
Method: single particle / : Pallesen J, Ozorowski G, de Val N, Ward AB

EMDB-8729: 
Cryo-EM reconstruction of the HIV-1 BG505 SOSIP.664 Env trimer in complex with soluble CD4 (D1-D2)
Method: single particle / : Lyumkis D, de Val N, Ward AB

EMDB-8730: 
Cryo-EM reconstruction of the HIV-1 BG505 SOSIP.664 Env trimer in complex with soluble CD4 (D1-D2) and fragment antigen binding variable domain of 17b
Method: single particle / : Lyumkis D, de Val N, Ward AB

EMDB-8713: 
Cryo-EM reconstruction of B41 SOSIP.664 in complex with soluble CD4 (D1-D2) and fragment antigen binding of 17b
Method: single particle / : Pallesen J, Ozorowski G, de Val N, Ward AB

EMDB-8717: 
Cryo-EM reconstruction of B41 SOSIP.664 in complex with fragment antigen binding of b12
Method: single particle / : Pallesen J, Ozorowski G, de Val N, Ward AB

PDB-5vn3: 
Cryo-EM model of B41 SOSIP.664 in complex with soluble CD4 (D1-D2) and fragment antigen binding variable domain of 17b
Method: single particle / : Ozorowski G, Pallesen J, Ward AB

PDB-5vn8: 
Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12
Method: single particle / : Ozorowski G, Pallesen J, Ward AB, Cottrell CA

EMDB-3059: 
ZM197 SOSIP.664 trimer in complex with VRC01 Fab
Method: single particle / : Lee JH, Ward AB

EMDB-2753: 
PGT124 in complex with BG505 SOSIP.664 Env trimer
Method: single particle / : Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien JP, Hua Y, Cupo A, Moore JP, Ward AB, Paulson JC, Burton DR, Wilson IA

EMDB-2853: 
3BNC117 Fab in complex with BG505 SOSIP.664 Trimer
Method: single particle / : Ozorowski G, Derking R, Sanders RW, Ward AB
EMDB-6249: 
1NC9 Fab in complex with BG505 SOSIP.664 Trimer
Method: single particle / : Ozorowski G, Derking R, Sanders RW, Ward AB

EMDB-6250: 
CH103 Fab in complex with BG505 SOSIP.664 Trimer
Method: single particle / : Ozorowski G, Derking R, Haynes B, Kwong PD, Sanders RW, Ward AB
EMDB-6251: 
CH106 Fab in complex with BG505 SOSIP.664 Trimer
Method: single particle / : Ozorowski G, Derking R, Sanders RW, Ward AB
EMDB-6252: 
VRC01 Fab in complex with BG505 SOSIP.664 Trimer
Method: single particle / : Torres JL, Ozorowski G, Derking R, Sanders RW, Ward AB
EMDB-6256: 
A native-like SOSIP.664 trimer based on a HIV-1 subtype B env gene
Method: single particle / : Pugach P, Ozorowski G

EMDB-5982: 
Structure of 2G12 (Fab)2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single Particle Electron Microscopy
Method: single particle / : Murin CD, Julien JP, Stanfield R, Khayat R, Sok D, Burton DR, Wilson IA, Ward AB

EMDB-5983: 
Structure of 2G12 (Fab)2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single Particle Electron Microscopy
Method: single particle / : Murin CD, Julien JP, Sok D, Stanfield R, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, Ward AB

EMDB-5918: 
Negative stain reconstruction of PGT151 Fab in complex with BG505 Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5919: 
Negative stain reconstruction of PGT151 Fab in complex with JR-FL Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
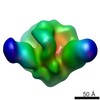
EMDB-5920: 
Negative stain reconstruction of PGT151 Fab in complex with cleaved IAVI C22 Env trimer containing the transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
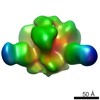
EMDB-5921: 
Negative stain reconstruction of PGT151 Fab in complex with the soluble Env trimer BG505 SOSIP
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5856: 
Negative stain reconstruction of HIV envelope glycoprotein BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab
Method: single particle / : Ward AB, Kim HJ

EMDB-5779: 
Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs
Method: single particle / : Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB

EMDB-5780: 
Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 2 PGV04 Fabs
Method: single particle / : Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB

EMDB-5781: 
Cryo-EM structure of the BG505 SOSIP.650 HIV-1 Env trimer with 3 PGV04 Fabs
Method: single particle / : Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB

EMDB-5782: 
Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer
Method: single particle / : Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB

PDB-3j5m: 
Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs
Method: single particle / : Lyumkis D, Julien JP, Wilson IA, Ward AB

EMDB-2427: 
Negative stain Electron Microscopy of BG505 SOSIP.664 gp140 in complex with PGV04
Method: single particle / : Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, Torrents A, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP

EMDB-5723: 
Negative stain Electron Microscopy of Bg505 SOSIP.664 in complex with sCD4 and 17b
Method: single particle / : Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, Torrents A, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP

EMDB-5700: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5701: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681.dV1V2V3
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5702: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681 in complex with Fab fragment of VRC PG04 monoclonal antibody
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5703: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681.dV1V2V3 in complex with Fab fragment of VRC PG04 monoclonal antibody
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5704: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.681 in complex with soluble CD4
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5705: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.664
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5706: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.664 in complex with Fab fragment of VRC PG04 monoclonal antibody
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB
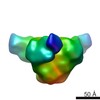
EMDB-5707: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.664.dV1V2V3 in complex with Fab fragment of VRC PG04 monoclonal antibody
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5708: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.664 in complex with soluble CD4
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5709: 
Negative stained image reconstruction of HIV KNH11444 subtype A SOSIP.664.dV1V2V3 in complex with soluble CD4
Method: single particle / : Khayat R, Lee JH, Wilson IA, Ward AB

EMDB-5624: 
Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans
Method: single particle / : Khayat R, Lee JH, Julien JP, Wilson IA, Ward AB

EMDB-2331: 
Negative stain reconstruction of BG505 SOSIP gp140 HIV-1 trimer in complex with PGT135 Fab
Method: single particle / : Lee JH, Kong L, Murin CD, Cupo A, Moore JP, Wilson IA, Ward AB
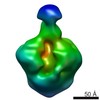
EMDB-2241: 
Negative stain reconstruction of PG9 Fab in complex with HIV-1 SOSIP gp140 trimer
Method: single particle / : Lee JH, Julien JP, Cupo A, Moore JP, Wilson IA, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
