[English] 日本語
 Yorodumi
Yorodumi- EMDB-8547: Kinesin-1 Binding Microtubules with ADP-AlFx in a One-head-bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8547 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Kinesin-1 Binding Microtubules with ADP-AlFx in a One-head-bound State (low-pass filtered to 8.5 angstrom) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Bos taurus (cattle) / Bos taurus (cattle) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.7 Å cryo EM / Resolution: 6.7 Å | |||||||||
 Authors Authors | Liu D / Liu X / Shang Z / Sindelar CV | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Structural basis of cooperativity in kinesin revealed by 3D reconstruction of a two-head-bound state on microtubules. Authors: Daifei Liu / Xueqi Liu / Zhiguo Shang / Charles V Sindelar /  Abstract: The detailed basis of walking by dimeric molecules of kinesin along microtubules has remained unclear, partly because available structural methods have been unable to capture microtubule-bound ...The detailed basis of walking by dimeric molecules of kinesin along microtubules has remained unclear, partly because available structural methods have been unable to capture microtubule-bound intermediates of this process. Utilizing novel electron cryomicroscopy methods, we solved structures of microtubule-attached, dimeric kinesin bound to an ATP analog. We find that under these conditions, the kinesin dimer can attach to the microtubule with either one or two motor domains, and we present sub-nanometer resolution reconstructions of both states. The former structure reveals a novel kinesin conformation that revises the current understanding of how ATP binding is coupled to forward stepping of the motor. The latter structure indicates how tension between the two motor domains keeps their cycles out of phase in order to stimulate directional motility. The methods presented here pave the way for future structural studies of a variety of challenging macromolecules that bind to microtubules and other filaments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8547.map.gz emd_8547.map.gz | 147.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8547-v30.xml emd-8547-v30.xml emd-8547.xml emd-8547.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8547.png emd_8547.png | 74.5 KB | ||
| Others |  emd_8547_half_map_1.map.gz emd_8547_half_map_1.map.gz emd_8547_half_map_2.map.gz emd_8547_half_map_2.map.gz | 118.5 MB 118.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8547 http://ftp.pdbj.org/pub/emdb/structures/EMD-8547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8547 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8547.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8547.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_8547_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_8547_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Kinesin-1 (1-420) on microtubules with ADP-AlFx in a one-head-bou...
| Entire | Name: Kinesin-1 (1-420) on microtubules with ADP-AlFx in a one-head-bound state |
|---|---|
| Components |
|
-Supramolecule #1: Kinesin-1 (1-420) on microtubules with ADP-AlFx in a one-head-bou...
| Supramolecule | Name: Kinesin-1 (1-420) on microtubules with ADP-AlFx in a one-head-bound state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Macromolecule #1: Kinesin-1
| Macromolecule | Name: Kinesin-1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MADLAESNIK VMSRFRPLNE SEVNRGDKYI AKFQGEDTVV IASKPYAFDR VFQSSTSQEQ VYNDSAKKIV KDVLEGYNGT IFAYGQTSSG KTHTMEGKLH DPEGMGIIPR IVQDIFNYIY SMDENLEFHI KVSYFEIYLD KIRDLLDVSK TNLSVHEDKN RVPYVKGSTE ...String: MADLAESNIK VMSRFRPLNE SEVNRGDKYI AKFQGEDTVV IASKPYAFDR VFQSSTSQEQ VYNDSAKKIV KDVLEGYNGT IFAYGQTSSG KTHTMEGKLH DPEGMGIIPR IVQDIFNYIY SMDENLEFHI KVSYFEIYLD KIRDLLDVSK TNLSVHEDKN RVPYVKGSTE RFVSSPDEVM DTIDEGKSNR HVAVTNMNEH SSRSHSIFLI NVKQENTQTE QKLSGKLYLV DLAGSEKVSK TGAEGAVLDE AKNINKSLSA LGNVISALAE GSTYVPYRDS KMTRILQDSL GGNSRTTIVI SSSPSSYNES ETKSTLLFGQ RAKTIKNTVS VNVELTAEQW KKKYEKEKEK NKILRNTIQW LENELNRWRN GETVPIDEQF DKEKANLEAF TVDKDITLTN DKPATAIGVI GNFTDAERRK |
-Macromolecule #2: Alpha-tubulin
| Macromolecule | Name: Alpha-tubulin / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHPE QLITGKEDAA NNYARGHYTI GKEIIDLVLD RIRKLADQCT GLQGFLVFHS FGGGTGSGFT SLLMERLSVD YGKKSKLEFS IYPAPQVSTA VVEPYNSILT THTTLEHSDC AFMVDNEAIY DICRRNLDIE RPTYTNLNRL ISQIVSSITA SLRFDGALNV DLTEFQTNLV PYPRIHFPLA TYAPVISAEK AYHEQLSVAE ITNACFEPAN QMVKCDPRHG KYMACCLLYR GDVVPKDVNA AIATIKTKRS IQFVDWCPTG FKVGINYQPP TVVPGGDLAK VQRAVCMLSN TTAIAEAWAR LDHKFDLMYA KRAFVHWYVG EGMEEGEFSE AREDMAALEK DYEEVGVDSV EGEGEEEGEE Y |
-Macromolecule #3: Beta-tubulin
| Macromolecule | Name: Beta-tubulin / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Sequence | String: MREIVHLQAG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGNYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDAVLDVV RKEAESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEFP DRIMNTFSVV ...String: MREIVHLQAG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGNYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDNF VFGQSGAGNN WAKGHYTEGA ELVDAVLDVV RKEAESCDCL QGFQLTHSLG GGTGSGMGTL LISKIREEFP DRIMNTFSVV PSPKVSDTVV EPYNATLSVH QLVENTDETY CIDNEALYDI CFRTLKLTTP TYGDLNHLVS ATMSGVTTCL RFPGQLNADL RKLAVNMVPF PRLHFFMPGF APLTSRGSQQ YRALTVPELT QQMFDAKNMM AACDPRHGRY LTVAAVFRGR MSMKEVDEQM LSVQSKNSSY FVEWIPNNVK TAVCDIPPRG LKMAATFIGN STAIQELFKR ISEQFTAMFR RKAFLHWYTG EGMDEMEFTE AESNMNDLVS EYQQYQDATA EEGEFEEEAE EEVA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average electron dose: 1.65 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: PROJECTION MATCHING |
|---|---|
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.7 Å / Resolution method: FSC 0.143 CUT-OFF Details: a modified version of frealign was used for reconstruction, kinesin dimer was masked before FSC calculation using half maps. Number images used: 44298 |
 Movie
Movie Controller
Controller







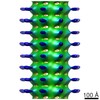



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































