[English] 日本語
 Yorodumi
Yorodumi- EMDB-6639: Cryo-electron microscopy of a deletion mutant (d385-399) of Flock... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6639 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy of a deletion mutant (d385-399) of Flock House virus VLP | |||||||||
 Map data Map data | Reconstruction of deletion mutant (d385-399) Flock House virus VLP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Flock House virus / VLP / Flock House virus / VLP /  gamma gamma | |||||||||
| Biological species |   Flock house virus Flock house virus | |||||||||
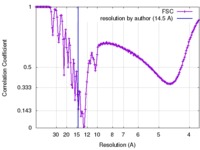
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 14.5 Å cryo EM / Resolution: 14.5 Å | |||||||||
 Authors Authors | Bajaj S / Dey D / Kumar M / Banerjee M | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2016 Journal: J Mol Biol / Year: 2016Title: Non-Enveloped Virus Entry: Structural Determinants and Mechanism of Functioning of a Viral Lytic Peptide. Authors: Saumya Bajaj / Debajit Dey / Rohan Bhukar / Mohit Kumar / Manidipa Banerjee /  Abstract: In the absence of lipid envelopes and associated fusion proteins, non-enveloped viruses employ membrane lytic peptides to breach the limiting membranes of host cells. Although several of these lytic ...In the absence of lipid envelopes and associated fusion proteins, non-enveloped viruses employ membrane lytic peptides to breach the limiting membranes of host cells. Although several of these lytic peptides have been identified and characterized, their manner of deployment and interaction with host membranes remains unclear in most cases. We are using the gamma peptide of Flock House Virus (FHV), a model non-enveloped virus, to understand the mechanistic details of non-enveloped virus interaction with host cell membranes. We utilized a combination of biophysical assays, molecular dynamics simulation studies, and single-particle cryo-electron microscopy to elucidate the functional and structural determinants for membrane penetration by gamma in context of the FHV capsid. Although the amphipathic, helical N-terminal region of gamma (γ1) was previously thought to be the membrane-penetrating module, with the C-terminal region having a supporting role in correct structural positioning of γ1, we demonstrate that the C terminus of gamma directly participates in membrane penetration. Our studies suggest that full-length gamma, including the hydrophobic C terminus, forms an alpha-helical hairpin motif, and any disruption in this motif drastically reduces its functionality, in spite of the correct positioning of amphipathic γ1 in the virus capsid. Taken together, our data suggest that the most effective module for membrane disruption is a pentameric unit of full-length gamma, released from the virus, which associates with membranes via both N- and C-terminal ends. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6639.map.gz emd_6639.map.gz | 47.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6639-v30.xml emd-6639-v30.xml emd-6639.xml emd-6639.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6639_fsc.xml emd_6639_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_6639.tif emd_6639.tif | 2.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6639 http://ftp.pdbj.org/pub/emdb/structures/EMD-6639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6639 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6639.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6639.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of deletion mutant (d385-399) Flock House virus VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Virus-like particle of deletion mutant (d385-399) of Flock House virus
| Entire | Name: Virus-like particle of deletion mutant (d385-399) of Flock House virus |
|---|---|
| Components |
|
-Supramolecule #1000: Virus-like particle of deletion mutant (d385-399) of Flock House virus
| Supramolecule | Name: Virus-like particle of deletion mutant (d385-399) of Flock House virus type: sample / ID: 1000 Oligomeric state: 180 molecules of capsid protein make up one particle Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 9 MDa |
-Supramolecule #1: Flock house virus
| Supramolecule | Name: Flock house virus / type: virus / ID: 1 Details: Residues 385-399 of the capsid protein have been deleted NCBI-ID: 12287 / Sci species name: Flock house virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Costelytra zealandica (beetle) / synonym: INVERTEBRATES Costelytra zealandica (beetle) / synonym: INVERTEBRATES |
| Host system | Organism:   Spodoptera frugiperda (fall armyworm) / Recombinant cell: Sf21 / Recombinant plasmid: pBacPAK9 Spodoptera frugiperda (fall armyworm) / Recombinant cell: Sf21 / Recombinant plasmid: pBacPAK9 |
| Molecular weight | Theoretical: 9 MDa |
| Virus shell | Shell ID: 1 / Name: Alpha / Diameter: 336 Å / T number (triangulation number): 3 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM HEPES, 5mM CaCl2 |
| Grid | Details: 300 mesh copper, Quantifoil R2/2 grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 77 K / Instrument: FEI VITROBOT MARK IV Method: Blot for 3 seconds at a blot force of 0 before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 1.2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 80000 Bright-field microscopy / Cs: 1.2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Temperature | Min: 77 K / Max: 97 K / Average: 94 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 80,000x magnification |
| Date | Jan 8, 2016 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Number real images: 1199 / Average electron dose: 10 e/Å2 / Bits/pixel: 2 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller