+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4153 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S.cerevisiae partial Elp123 sub-complex | |||||||||
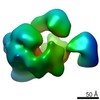
 Map data Map data | Partial Elp123 sub-complex from S.cerevisiae in which one of the Elp2 subunits is missing. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 31.0 Å negative staining / Resolution: 31.0 Å | |||||||||
 Authors Authors | Dauden MI / Kosinski J / Kolaj-Robin O / Desfosses A / Ori A / Faux C / Hoffmann NA / Onuma OF / Breuring KD / Beck M ...Dauden MI / Kosinski J / Kolaj-Robin O / Desfosses A / Ori A / Faux C / Hoffmann NA / Onuma OF / Breuring KD / Beck M / Sachse C / Seraphin B / Glatt S / Mueller CW | |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2017 Journal: EMBO Rep / Year: 2017Title: Architecture of the yeast Elongator complex. Authors: Maria I Dauden / Jan Kosinski / Olga Kolaj-Robin / Ambroise Desfosses / Alessandro Ori / Celine Faux / Niklas A Hoffmann / Osita F Onuma / Karin D Breunig / Martin Beck / Carsten Sachse / ...Authors: Maria I Dauden / Jan Kosinski / Olga Kolaj-Robin / Ambroise Desfosses / Alessandro Ori / Celine Faux / Niklas A Hoffmann / Osita F Onuma / Karin D Breunig / Martin Beck / Carsten Sachse / Bertrand Séraphin / Sebastian Glatt / Christoph W Müller /    Abstract: The highly conserved eukaryotic Elongator complex performs specific chemical modifications on wobble base uridines of tRNAs, which are essential for proteome stability and homeostasis. The complex is ...The highly conserved eukaryotic Elongator complex performs specific chemical modifications on wobble base uridines of tRNAs, which are essential for proteome stability and homeostasis. The complex is formed by six individual subunits (Elp1-6) that are all equally important for its tRNA modification activity. However, its overall architecture and the detailed reaction mechanism remain elusive. Here, we report the structures of the fully assembled yeast Elongator and the Elp123 sub-complex solved by an integrative structure determination approach showing that two copies of the Elp1, Elp2, and Elp3 subunits form a two-lobed scaffold, which binds Elp456 asymmetrically. Our topological models are consistent with previous studies on individual subunits and further validated by complementary biochemical analyses. Our study provides a structural framework on how the tRNA modification activity is carried out by Elongator. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4153.map.gz emd_4153.map.gz | 33.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4153-v30.xml emd-4153-v30.xml emd-4153.xml emd-4153.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4153.png emd_4153.png | 147.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4153 http://ftp.pdbj.org/pub/emdb/structures/EMD-4153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4153 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4153.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4153.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Partial Elp123 sub-complex from S.cerevisiae in which one of the Elp2 subunits is missing. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S.cerevisiae partial Elp123 sub-complex, without one of the Elp2 ...
| Entire | Name: S.cerevisiae partial Elp123 sub-complex, without one of the Elp2 subunits. |
|---|---|
| Components |
|
-Supramolecule #1: S.cerevisiae partial Elp123 sub-complex, without one of the Elp2 ...
| Supramolecule | Name: S.cerevisiae partial Elp123 sub-complex, without one of the Elp2 subunits. type: complex / ID: 1 / Parent: 0 Details: Composed of two copies of Elp1 and Elp3 and only one copy of Elp2. |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Strain: BS1173 Saccharomyces cerevisiae (brewer's yeast) / Strain: BS1173 |
| Molecular weight | Theoretical: 530 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.035 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: 3.5 ul aliquots of freshly purified Elp123 complex were applied to glow discharged carbon copper-collodion (Sigma) grids for 2 min and stained with a 1% uranyl acetate solution (w/v) | ||||||||||||
| Grid | Model: Plano D-35578 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Details: 0.45mBar, 20mA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 6.3 mm / Nominal magnification: 49000 Bright-field microscopy / Cs: 6.3 mm / Nominal magnification: 49000 |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number grids imaged: 1 / Number real images: 216 / Average electron dose: 16.0 e/Å2 |
- Image processing
Image processing
| Particle selection | Number selected: 50034 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 3) |
| Startup model | Type of model: RANDOM CONICAL TILT / Random conical tilt - Number images: 120 / Random conical tilt - Tilt angle: 55 degrees |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: Xmipp |
| Final 3D classification | Number classes: 2 / Avg.num./class: 3200 / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 31.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 1946 |
 Movie
Movie Controller
Controller