[English] 日本語
 Yorodumi
Yorodumi- EMDB-4128: Binding of the C-terminal GQYL motif of the bacterial proteasome ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4128 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defenses / proteasome accessory complex / positive regulation of proteasomal protein catabolic process /  zymogen binding / zymogen binding /  proteasome binding / proteasome binding /  proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process ...symbiont-mediated perturbation of host defenses / proteasome accessory complex / positive regulation of proteasomal protein catabolic process / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process ...symbiont-mediated perturbation of host defenses / proteasome accessory complex / positive regulation of proteasomal protein catabolic process /  zymogen binding / zymogen binding /  proteasome binding / proteasome binding /  proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / peptidoglycan-based cell wall / proteasomal protein catabolic process / modification-dependent protein catabolic process / protein homooligomerization / extracellular region / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / peptidoglycan-based cell wall / proteasomal protein catabolic process / modification-dependent protein catabolic process / protein homooligomerization / extracellular region /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Bolten M / Delley CL / Leibundgut M / Boehringer D / Ban N / Weber-Ban E | |||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome. Authors: Marcel Bolten / Cyrille L Delley / Marc Leibundgut / Daniel Boehringer / Nenad Ban / Eilika Weber-Ban /  Abstract: Mycobacterium tuberculosis harbors proteasomes that recruit substrates for degradation through an ubiquitin-like modification pathway. Recently, a non-ATPase activator termed Bpa (bacterial ...Mycobacterium tuberculosis harbors proteasomes that recruit substrates for degradation through an ubiquitin-like modification pathway. Recently, a non-ATPase activator termed Bpa (bacterial proteasome activator) was shown to support an alternate proteasomal degradation pathway. Here, we present the cryo-electron microscopy (cryo-EM) structure of Bpa in complex with the 20S core particle (CP). For docking into the cryo-EM density, we solved the X-ray structure of Bpa, showing that it forms tight four-helix bundles arranged into a 12-membered ring with a 40 Å wide central pore and the C-terminal helix of each protomer protruding from the ring. The Bpa model was fitted into the cryo-EM map of the Bpa-CP complex, revealing its architecture and striking symmetry mismatch. The Bpa-CP interface was resolved to 3.5 Å, showing the interactions between the C-terminal GQYL motif of Bpa and the proteasome α-rings. This docking mode is related to the one observed for eukaryotic activators with features specific to the bacterial complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4128.map.gz emd_4128.map.gz | 201.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4128-v30.xml emd-4128-v30.xml emd-4128.xml emd-4128.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
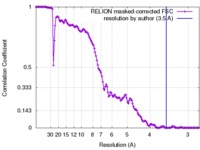
| FSC (resolution estimation) |  emd_4128_fsc.xml emd_4128_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4128.png emd_4128.png | 163.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4128 http://ftp.pdbj.org/pub/emdb/structures/EMD-4128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4128 | HTTPS FTP |
-Related structure data
| Related structure data |  5lzpMC  4127C  5lfjC  5lfpC  5lfqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4128.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4128.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : proteasome in complex with bacterial proteasome activator
| Entire | Name: proteasome in complex with bacterial proteasome activator |
|---|---|
| Components |
|
-Supramolecule #1: proteasome in complex with bacterial proteasome activator
| Supramolecule | Name: proteasome in complex with bacterial proteasome activator type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) / Location in cell: cytoplasm Mycobacterium tuberculosis H37Rv (bacteria) / Location in cell: cytoplasm |
| Molecular weight | Theoretical: 930 KDa |
-Macromolecule #1: Proteasome subunit alpha
| Macromolecule | Name: Proteasome subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number:  proteasome endopeptidase complex proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 26.024971 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SPEQAMRERS ELARKGIARA KSVVALAYAG GVLFVAENPS RSLQKISELY DRVGFAAAGK FNEFDNLRRG GIQFADTRGY AYDRRDVTG RQLANVYAQT LGTIFTEQAK PYEVELCVAE VAHYGETKRP ELYRITYDGS IADEPHFVVM GGTTEPIANA L KESYAENA ...String: SPEQAMRERS ELARKGIARA KSVVALAYAG GVLFVAENPS RSLQKISELY DRVGFAAAGK FNEFDNLRRG GIQFADTRGY AYDRRDVTG RQLANVYAQT LGTIFTEQAK PYEVELCVAE VAHYGETKRP ELYRITYDGS IADEPHFVVM GGTTEPIANA L KESYAENA SLTDALRIAV AALRAGSADT SGGDQPTLGV ASLEVAVLDA NRPRRAFRRI TGSALQALLV DQESPQSDGE SS G |
-Macromolecule #2: Bacterial proteasome activator
| Macromolecule | Name: Bacterial proteasome activator / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 19.792113 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MHHHHHHVIG LSTGSDDDDV EVIGGVDPRL IAVQENDSDE SSLTDLVEQP AKVMRIGTMI KQLLEEVRAA PLDEASRNRL RDIHATSIR ELEDGLAPEL REELDRLTLP FNEDAVPSDA ELRIAQAQLV GWLEGLFHGI QTALFAQQMA ARAQLQQMRQ G ALPPGVGK SGQHGHGTGQ YL |
-Macromolecule #3: Proteasome subunit beta
| Macromolecule | Name: Proteasome subunit beta / type: protein_or_peptide / ID: 3 / Number of copies: 14 / Enantiomer: LEVO / EC number:  proteasome endopeptidase complex proteasome endopeptidase complex |
|---|---|
| Source (natural) | Organism:   Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 25.457504 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: ATIVALKYPG GVVMAGDRRS TQGNMISGRD VRKVYITDDY TATGIAGTAA VAVEFARLYA VELEHYEKLE GVPLTFAGKI NRLAIMVRG NLAAAMQGLL ALPLLAGYDI HASDPQSAGR IVSFDAAGGW NIEEEGYQAV GSGSLFAKSS MKKLYSQVTD G DSGLRVAV ...String: ATIVALKYPG GVVMAGDRRS TQGNMISGRD VRKVYITDDY TATGIAGTAA VAVEFARLYA VELEHYEKLE GVPLTFAGKI NRLAIMVRG NLAAAMQGLL ALPLLAGYDI HASDPQSAGR IVSFDAAGGW NIEEEGYQAV GSGSLFAKSS MKKLYSQVTD G DSGLRVAV EALYDAADDD SATGGPDLVR GIFPTAVIID ADGAVDVPES RIAELARAII ESRSGADTFG SDGGEKWSHP QF EK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.102 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER Details: Quantifoil R 2/2 with an additional thin carbon layer | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 280.5 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 100000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Average electron dose: 25.0 e/Å2 Details: Drift corrected in post-processing. 4 images per hole. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5lzp: |
 Movie
Movie Controller
Controller