[English] 日本語
 Yorodumi
Yorodumi- EMDB-3312: HIV-1 cleaved wild type JR-FL EnvdCT trimer in complex with PGT15... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3312 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 and 10E8 Fabs at 8.8 A resolution | |||||||||
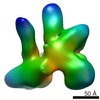
 Map data Map data | Reconstruction of EnvdCT in complex with 10E8 and PGT151 Fabs. Partial density of a third 10E8 Fab visible due to partial binding occupancy. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV-1 / Env / PGT151 / 10E8 / HIV-1 / Env / PGT151 / 10E8 /  antibody / MPER antibody / MPER | |||||||||
| Biological species |    Human Immunodeficiency Virus-1 / Human Immunodeficiency Virus-1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 8.8 Å cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Lee JH / Ward AB | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Authors: Jeong Hyun Lee / Gabriel Ozorowski / Andrew B Ward /  Abstract: The envelope glycoprotein trimer (Env) on the surface of HIV-1 recognizes CD4(+) T cells and mediates viral entry. During this process, Env undergoes substantial conformational rearrangements, making ...The envelope glycoprotein trimer (Env) on the surface of HIV-1 recognizes CD4(+) T cells and mediates viral entry. During this process, Env undergoes substantial conformational rearrangements, making it difficult to study in its native state. Soluble stabilized trimers have provided valuable insights into the Env structure, but they lack the hydrophobic membrane proximal external region (MPER, an important target of broadly neutralizing antibodies), the transmembrane domain, and the cytoplasmic tail. Here we present (i) a cryogenic electron microscopy (cryo-EM) structure of a clade B virus Env, which lacks only the cytoplasmic tail and is stabilized by the broadly neutralizing antibody PGT151, at a resolution of 4.2 angstroms and (ii) a reconstruction of this form of Env in complex with PGT151 and MPER-targeting antibody 10E8 at a resolution of 8.8 angstroms. These structures provide new insights into the wild-type Env structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3312.map.gz emd_3312.map.gz | 58.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3312-v30.xml emd-3312-v30.xml emd-3312.xml emd-3312.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3312.png emd_3312.png | 565.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3312 http://ftp.pdbj.org/pub/emdb/structures/EMD-3312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3312 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3312 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3312.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3312.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of EnvdCT in complex with 10E8 and PGT151 Fabs. Partial density of a third 10E8 Fab visible due to partial binding occupancy. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cleaved JR-FL EnvdCT in complex with PGT151 and 10E8 Fabs
| Entire | Name: Cleaved JR-FL EnvdCT in complex with PGT151 and 10E8 Fabs |
|---|---|
| Components |
|
-Supramolecule #1000: Cleaved JR-FL EnvdCT in complex with PGT151 and 10E8 Fabs
| Supramolecule | Name: Cleaved JR-FL EnvdCT in complex with PGT151 and 10E8 Fabs type: sample / ID: 1000 Oligomeric state: One trimer bound to one PGT151 and two 10E8 Fabs Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 585 KDa |
-Macromolecule #1: HIV-1 Envelope glycoprotein
| Macromolecule | Name: HIV-1 Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: HIV-1 Env Details: wild-type JR-FL Env trimer with the cytoplasmic tail truncated Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human Immunodeficiency Virus-1 / Strain: JR-FL / synonym: HIV-1 Human Immunodeficiency Virus-1 / Strain: JR-FL / synonym: HIV-1 |
| Molecular weight | Theoretical: 435 KDa |
| Recombinant expression | Organism:  Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F / Recombinant plasmid: phCMV3 Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F / Recombinant plasmid: phCMV3 |
-Macromolecule #2: Immunoglobulin G PGT151
| Macromolecule | Name: Immunoglobulin G PGT151 / type: protein_or_peptide / ID: 2 / Name.synonym: IgG PGT151 / Details: PGT151 cleaved into Fab / Number of copies: 1 / Oligomeric state: Heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F |
-Macromolecule #3: Immunoglobulin G 10E8
| Macromolecule | Name: Immunoglobulin G 10E8 / type: protein_or_peptide / ID: 3 / Name.synonym: IgG 10E8 / Details: 10E8 expressed as Fab / Number of copies: 2 / Oligomeric state: Heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 50 MDa |
| Recombinant expression | Organism:  Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F Mammalian (mammals) / Recombinant strain: Human / Recombinant cell: HEK293F |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Tris pH 7.4, 150 mM NaCl, 0.1% DDM, 0.03 mg/mL sodium deoxycholate |
| Grid | Details: 400 mesh C-Flat, CF-2/2-4C, plasma cleaned for 5 seconds |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Samples were treated with biobeads prior to freezing. Method: Grids were manually plunged at RT. |
- Electron microscopy #1
Electron microscopy #1
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 22500 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Microscopy ID | 1 |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism corrected at 22,500x magnification. |
| Date | Dec 16, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Sampling interval: 5.0 µm / Number real images: 3969 / Average electron dose: 32.4 e/Å2 Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. However, in the Nov session, the microscope experienced FEG instability resulting in ...Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. However, in the Nov session, the microscope experienced FEG instability resulting in intensity drop over the course of data collection. |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #2
Electron microscopy #2
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 22500 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Microscopy ID | 2 |
| Alignment procedure | Legacy - Astigmatism: Objective astigmatism corrected at 22,500x magnification. |
| Details | Unstable beam intensity over the course of data collection. |
| Date | Nov 17, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Sampling interval: 5.0 µm / Number real images: 3969 / Average electron dose: 28.1 e/Å2 Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. However, in the Nov session, the microscope experienced FEG instability resulting in ...Details: Each full dose image is an aligned stack of frames recorded each using a dose of ~10 e-/Angstrom^2/sec. However, in the Nov session, the microscope experienced FEG instability resulting in intensity drop over the course of data collection. |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.8 Å / Resolution method: OTHER / Software - Name: Relion / Number images used: 15525 |
| Details | The full data set was sorted into multiple 3D classes, which all had various Fab binding stoichiometries of PGT151 and 10E8. This reconstruction is the only sub-population that refined below 10A resolution. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: L / Chain - #1 - Chain ID: H / Chain - #2 - Chain ID: P |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The N-terminal helix of the gp41 peptide and Fab constant regions were removed prior to fitting. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller