[English] 日本語
 Yorodumi
Yorodumi- EMDB-1392: A three-dimensional cryo-electron microscopy structure of the bac... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1392 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head. | |||||||||
 Map data Map data | cryo-EM reconstruction of the bacteriophage phiKZ capsid | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 18.0 Å cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Fokine A / Kostyuchenko V / Efimov A / Kurochkina L / Sykilinda N / Robben J / Volckaert G / Hoenger A / Chipman P / Battisti A ...Fokine A / Kostyuchenko V / Efimov A / Kurochkina L / Sykilinda N / Robben J / Volckaert G / Hoenger A / Chipman P / Battisti A / Rossmann M / Mesyanzhinov V | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2005 Journal: J Mol Biol / Year: 2005Title: A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head. Authors: Andrei Fokine / Victor A Kostyuchenko / Andrey V Efimov / Lidia P Kurochkina / Nina N Sykilinda / Johan Robben / Guido Volckaert / Andreas Hoenger / Paul R Chipman / Anthony J Battisti / ...Authors: Andrei Fokine / Victor A Kostyuchenko / Andrey V Efimov / Lidia P Kurochkina / Nina N Sykilinda / Johan Robben / Guido Volckaert / Andreas Hoenger / Paul R Chipman / Anthony J Battisti / Michael G Rossmann / Vadim V Mesyanzhinov /  Abstract: The three-dimensional structure of the Pseudomonas aeruginosa bacteriophage phiKZ head has been determined by cryo-electron microscopy and image reconstruction to 18A resolution. The head has ...The three-dimensional structure of the Pseudomonas aeruginosa bacteriophage phiKZ head has been determined by cryo-electron microscopy and image reconstruction to 18A resolution. The head has icosahedral symmetry measuring 1455 A in diameter along 5-fold axes and a unique portal vertex to which is attached an approximately 1800 A-long contractile tail. The 65 kDa major capsid protein, gp120, is organized into a surface lattice of hexamers, with T = 27 triangulation. The shape and size of the hexamers is similar to the hexameric building blocks of the bacteriophages T4, phi29, P22, and HK97. Pentameric vertices of the capsid are occupied by complexes composed of several special vertex proteins. The double-stranded genomic DNA is packaged into a highly condensed series of layers, separated by 24 A, that follow the contour of the inner wall of the capsid. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1392.map.gz emd_1392.map.gz | 77.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1392-v30.xml emd-1392-v30.xml emd-1392.xml emd-1392.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1392.gif 1392.gif emd_1392.tif emd_1392.tif | 117.8 KB 1.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1392 http://ftp.pdbj.org/pub/emdb/structures/EMD-1392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1392 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1392.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1392.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM reconstruction of the bacteriophage phiKZ capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.242 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
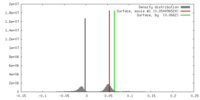
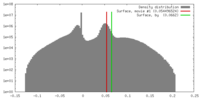
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bacteriophage phiKZ capsid
| Entire | Name: Bacteriophage phiKZ capsid |
|---|---|
| Components |
|
-Supramolecule #1000: Bacteriophage phiKZ capsid
| Supramolecule | Name: Bacteriophage phiKZ capsid / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Pseudomonas phage phiKZ
| Supramolecule | Name: Pseudomonas phage phiKZ / type: virus / ID: 1 / Name.synonym: phage phiKZ head, bacteriophage phiKZ / NCBI-ID: 169683 / Sci species name: Pseudomonas phage phiKZ / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: phage phiKZ head, bacteriophage phiKZ |
|---|---|
| Host (natural) | Organism:   Pseudomonas aeruginosa (bacteria) / synonym: BACTERIA(EUBACTERIA) Pseudomonas aeruginosa (bacteria) / synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Name: gp120 / Diameter: 1455 Å / T number (triangulation number): 27 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM tris-Acetate-EDTA buffer (pH 7.5) |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 40 % / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: in house manufactured / Method: hand blot 3 seconds, pluging during blot |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 33200 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 33000 Bright-field microscopy / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Alignment procedure | Legacy - Astigmatism: live fft |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 92 / Average electron dose: 20 e/Å2 / Bits/pixel: 8 |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final angle assignment | Details: SPIDER theta 37 degrees phi 72 degrees |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: SPIDER ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: SPIDERDetails: reconstruction was performed using the icosahedral symmetry Number images used: 2000 |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)