[English] 日本語
 Yorodumi
Yorodumi- EMDB-8075: Structure of the cellulose synthase complex of Gluconacetobacter ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8075 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the cellulose synthase complex of Gluconacetobacter hansenii at 23.4 Angstrom resolution | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Gluconacetobacter hansenii ATCC 23769 (bacteria) / Gluconacetobacter hansenii ATCC 23769 (bacteria) /  Komagataeibacter hansenii (bacteria) Komagataeibacter hansenii (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 23.4 Å negative staining / Resolution: 23.4 Å | |||||||||
 Authors Authors | Nixon BT / Du J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
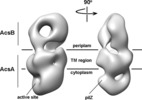
 Citation Citation |  Journal: PLoS One / Year: 2016 Journal: PLoS One / Year: 2016Title: Structure of the Cellulose Synthase Complex of Gluconacetobacter hansenii at 23.4 Å Resolution. Authors: Juan Du / Venkata Vepachedu / Sung Hyun Cho / Manish Kumar / B Tracy Nixon /  Abstract: Bacterial crystalline cellulose is used in biomedical and industrial applications, but the molecular mechanisms of synthesis are unclear. Unlike most bacteria, which make non-crystalline cellulose, ...Bacterial crystalline cellulose is used in biomedical and industrial applications, but the molecular mechanisms of synthesis are unclear. Unlike most bacteria, which make non-crystalline cellulose, Gluconacetobacter hansenii extrudes profuse amounts of crystalline cellulose. Its cellulose synthase (AcsA) exists as a complex with accessory protein AcsB, forming a 'terminal complex' (TC) that has been visualized by freeze-fracture TEM at the base of ribbons of crystalline cellulose. The catalytic AcsAB complex is embedded in the cytoplasmic membrane. The C-terminal portion of AcsC is predicted to form a translocation channel in the outer membrane, with the rest of AcsC possibly interacting with AcsD in the periplasm. It is thus believed that synthesis from an organized array of TCs coordinated with extrusion by AcsC and AcsD enable this bacterium to make crystalline cellulose. The only structural data that exist for this system are the above mentioned freeze-fracture TEM images, fluorescence microscopy images revealing that TCs align in a row, a crystal structure of AcsD bound to cellopentaose, and a crystal structure of PilZ domain of AcsA. Here we advance our understanding of the structural basis for crystalline cellulose production by bacterial cellulose synthase by determining a negative stain structure resolved to 23.4 Å for highly purified AcsAB complex that catalyzed incorporation of UDP-glucose into β-1,4-glucan chains, and responded to the presence of allosteric activator cyclic diguanylate. Although the AcsAB complex was functional in vitro, the synthesized cellulose was not visible in TEM. The negative stain structure revealed that AcsAB is very similar to that of the BcsAB synthase of Rhodobacter sphaeroides, a non-crystalline cellulose producing bacterium. The results indicate that the crystalline cellulose producing and non-crystalline cellulose producing bacteria share conserved catalytic and membrane translocation components, and support the hypothesis that it is the extrusion mechanism and order in linearly arrayed TCs that enables production of crystalline cellulose. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8075.map.gz emd_8075.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8075-v30.xml emd-8075-v30.xml emd-8075.xml emd-8075.xml | 10.3 KB 10.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8075.png emd_8075.png | 67.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8075 http://ftp.pdbj.org/pub/emdb/structures/EMD-8075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8075 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8075.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8075.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : membrane protein AcsAB
| Entire | Name: membrane protein AcsAB Biological membrane Biological membrane |
|---|---|
| Components |
|
-Supramolecule #1: membrane protein AcsAB
| Supramolecule | Name: membrane protein AcsAB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Gluconacetobacter hansenii ATCC 23769 (bacteria) Gluconacetobacter hansenii ATCC 23769 (bacteria) |
| Recombinant expression | Organism:  Komagataeibacter hansenii ATCC 23769 (bacteria) / Recombinant plasmid: pUCD2CDHisAcsAB Komagataeibacter hansenii ATCC 23769 (bacteria) / Recombinant plasmid: pUCD2CDHisAcsAB |
| Molecular weight | Theoretical: 170 KDa |
-Macromolecule #1: AcsAB Cellulose Synthase of Gluconacetobacter hansenii
| Macromolecule | Name: AcsAB Cellulose Synthase of Gluconacetobacter hansenii type: protein_or_peptide / ID: 1 Details: Heterodimer of AcsA and AcsB; membrane protein expressed as a fusion protein and then proteolytically processed. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataeibacter hansenii (bacteria) Komagataeibacter hansenii (bacteria) |
| Recombinant expression | Organism:  Komagataeibacter hansenii (bacteria) Komagataeibacter hansenii (bacteria) |
| Sequence | String: MPEVRSSTQS ESGMSQWMGK ILSIRGAGLI IGVFGLCALI AATSVTLPPE QQLIVAFVCV VIFFIVGHKP SRRSQIFLEV LSGLVSLRYL TWRLTETLSF DTWLQGLLGT MLLVAELYAL MMLFLSYFQT IAPLHRAPLP LPPNPDEWPT VDIFVPTYNE ELSIVRLTVL ...String: MPEVRSSTQS ESGMSQWMGK ILSIRGAGLI IGVFGLCALI AATSVTLPPE QQLIVAFVCV VIFFIVGHKP SRRSQIFLEV LSGLVSLRYL TWRLTETLSF DTWLQGLLGT MLLVAELYAL MMLFLSYFQT IAPLHRAPLP LPPNPDEWPT VDIFVPTYNE ELSIVRLTVL GSLGIDWPPE KVRVHILDDG RRPEFAAFAA ECGANYIARP TNEHAKAGNL NYAIGHTDGD YILIFDCDHV PTRAFLQLTM GWMVEDPKIA LMQTPHHFYS PDPFQRNLSA GYRTPPEGNL FYGVVQDGND FWDATFFCGS CAILRRTAIE QIGGFATQTV TEDAHTALKM QRLGWSTAYL RIPLAGGLAT ERLILHIGQR VRWARGMLQI FRIDNPLFGR GLSWGQRLCY LSAMTSFLFA VPRVIFLSSP LAFLFFGQNI IAASPLALLA YAIPHMFHAV GTASKINKGW RYSFWSEVYE TTMALFLVRV TIVTLLSPSR GKFNVTDKGG LLEKGYFDLG AVYPNIILGL IMFGGLARGV YELSFGHLDQ IAERAYLLNS AWAMLSLIII LAAIAVGRET QQKRNSHRIP ATIPVEVANA DGSIIVTGVT EDLSMGGAAV KMSWPAKLSG PTPVYIRTVL DGEELILPAR IIRAGNGRGI FIWTIDNLQQ EFSVIRLVFG RADAWVDWGN YKADRPLLSL MDMVLSVKGL FRSSGDIVHR SSPTKPSAGN ALSDDTNNPS RKERVLKGTV KMVSLLALLT FASSAQAASA PRAVAAKAPA HQPEASDLPP LPALLPATSG AAQAGSGDAG ANGPGSPTGQ PLAADSADAL VENAENTSDT ATVHNYTLKD LGAAGSITMR GLAPLQGIEF GIPSDQLVTS ARLVLSGSMS PNLRPETNSV TMTLNEQYIG TLRPDPAHPT FGPMSFEINP IFFVSGNRLN FNFASGSKGC SDITNDTLWA TISQNSQLQI TTIALPPRRL LSRLPQPFYD KNVRQHVTVP MVLAQTYDPQ ILKSAGILAS WFGKQTDFLG VTFPVSSTIP QSGNAILIGV ADELPTSLGR PQVNGPAVLE LPNPSDANAT ILVVTGRDRD EVITASKGIA FASAPLPTDS HMDVAPVDIA PRKPNDAPSF IAMDHPVRFG DLVTASKLQG TGFTSGVLSV PFRIPPDLYT WRNRPYKMQV RFRSPAGEAK DVEKSRLDVG INEVYLHSYP LRETHGLIGA VLQGVGLARP ASGMQVHDLD VPPWTVFGQD QLNFYFDAMP LARGICQSGA ANNAFHLGLD PDSTIDFSRA HHIAQMPNLA YMATVGFPFT TYADLSQTAV VLPEHPNAAT VGAYLDLMGF MGAATWYPVA GVDIVSADHV SDVADRNLLV ISTLATSGEI APLLSRSSYE VADGHLRTVS HASALDNAIK AVDDPLTAFR DRDSKPQDVD TPLTGGVGAM IEAESPLTAG RTVLALLSSD GAGLNNLLQM LGERKKQANI QGDLVVAHGE DLSSYRTSPV YTIGTLPLWL WPDWYMHNRP VRVLLVGLLG CILIVSVLAR ALARHATRRF KQLEDERRKS |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Staining | Type: NEGATIVE / Material: uranyl formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: NOT APPLICABLE |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 23.4 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 3168 |
 Movie
Movie Controller
Controller