[English] 日本語
 Yorodumi
Yorodumi- EMDB-6527: Negative stain EM reconstruction of Bundibugyo virus (BDBV) glyco... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6527 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
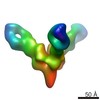
| Title | Negative stain EM reconstruction of Bundibugyo virus (BDBV) glycoprotein in complex with a human IgG1 Fab | |||||||||
 Map data Map data | Negative stain EM reconstruction of Bundibugyio virus (BDBV) mucin-deleted glycoprotein (GPdMuc) in complex with a human IgG1 Fab BDBV335 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Ebola virus / Ebola virus /  EBOV / GP / EBOV / GP /  antibody / antibody /  Bundibugyo virus / Bundibugyo virus /  BDBV / BDBV /  Fab Fab | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Ebola virus sp. Ebola virus sp. | |||||||||
| Method |  single particle reconstruction / Resolution: 17.0 Å single particle reconstruction / Resolution: 17.0 Å | |||||||||
 Authors Authors | Flyak AI / Shen X / Murin CD / Turner HL / Fusco ML / Lampley R / Kose N / Ilinykh PA / Kuzmina N / Branchizio A ...Flyak AI / Shen X / Murin CD / Turner HL / Fusco ML / Lampley R / Kose N / Ilinykh PA / Kuzmina N / Branchizio A / King H / Brown L / Davidson E / Doranz BJ / Slaughter JC / Sapparapu G / Klages C / Ksiazek TG / Saphire EO / Ward AB / Bukreyev A / Crowe JE | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Authors: Andrew I Flyak / Xiaoli Shen / Charles D Murin / Hannah L Turner / Joshua A David / Marnie L Fusco / Rebecca Lampley / Nurgun Kose / Philipp A Ilinykh / Natalia Kuzmina / Andre Branchizio / ...Authors: Andrew I Flyak / Xiaoli Shen / Charles D Murin / Hannah L Turner / Joshua A David / Marnie L Fusco / Rebecca Lampley / Nurgun Kose / Philipp A Ilinykh / Natalia Kuzmina / Andre Branchizio / Hannah King / Leland Brown / Christopher Bryan / Edgar Davidson / Benjamin J Doranz / James C Slaughter / Gopal Sapparapu / Curtis Klages / Thomas G Ksiazek / Erica Ollmann Saphire / Andrew B Ward / Alexander Bukreyev / James E Crowe /  Abstract: Recent studies have suggested that antibody-mediated protection against the Ebolaviruses may be achievable, but little is known about whether or not antibodies can confer cross-reactive protection ...Recent studies have suggested that antibody-mediated protection against the Ebolaviruses may be achievable, but little is known about whether or not antibodies can confer cross-reactive protection against viruses belonging to diverse Ebolavirus species, such as Ebola virus (EBOV), Sudan virus (SUDV), and Bundibugyo virus (BDBV). We isolated a large panel of human monoclonal antibodies (mAbs) against BDBV glycoprotein (GP) using peripheral blood B cells from survivors of the 2007 BDBV outbreak in Uganda. We determined that a large proportion of mAbs with potent neutralizing activity against BDBV bind to the glycan cap and recognize diverse epitopes within this major antigenic site. We identified several glycan cap-specific mAbs that neutralized multiple ebolaviruses, including SUDV, and a cross-reactive mAb that completely protected guinea pigs from the lethal challenge with heterologous EBOV. Our results provide a roadmap to develop a single antibody-based treatment effective against multiple Ebolavirus infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6527.map.gz emd_6527.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6527-v30.xml emd-6527-v30.xml emd-6527.xml emd-6527.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6527.jpg emd_6527.jpg | 439.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6527 http://ftp.pdbj.org/pub/emdb/structures/EMD-6527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6527 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6527.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6527.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM reconstruction of Bundibugyio virus (BDBV) mucin-deleted glycoprotein (GPdMuc) in complex with a human IgG1 Fab BDBV335 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fab fragment of BDBV335 human IgG1 monoclonal antibody to Bundibu...
| Entire | Name: Fab fragment of BDBV335 human IgG1 monoclonal antibody to Bundibugyo virus in complex with mucin-deleted Bundibugyo virus GP |
|---|---|
| Components |
|
-Supramolecule #1000: Fab fragment of BDBV335 human IgG1 monoclonal antibody to Bundibu...
| Supramolecule | Name: Fab fragment of BDBV335 human IgG1 monoclonal antibody to Bundibugyo virus in complex with mucin-deleted Bundibugyo virus GP type: sample / ID: 1000 Oligomeric state: Three Fabs (each consisting of one heavy chain and one light chain) bind to a single GP (consisting of a trimer of heterodimers of GP1 and GP2) Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 310 KDa |
-Macromolecule #1: human IgG1 monoclonal antibody Fab
| Macromolecule | Name: human IgG1 monoclonal antibody Fab / type: protein_or_peptide / ID: 1 / Name.synonym: Fab Details: Fabs were generated by optimized papain proteolysis of IgG1. There are three Fabs bound to each trimeric GP. Number of copies: 3 Oligomeric state: heterodimer (one heavy chain, one light chain) Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human / Cell: Peripheral blood mononuclear cells Homo sapiens (human) / synonym: Human / Cell: Peripheral blood mononuclear cells |
| Molecular weight | Theoretical: 50 KDa |
-Macromolecule #2: Bundibugyo virus glycoprotein, mucin-deleted
| Macromolecule | Name: Bundibugyo virus glycoprotein, mucin-deleted / type: protein_or_peptide / ID: 2 / Name.synonym: BDBV GPdMuc Details: Recombinantly expressed ectodomain of Bundibugyo virus GP. Mucin-like domains were deleted from the expression construct, as were the transmembrane domains and cytoplasmic tail. Number of copies: 1 Oligomeric state: trimer of heterodimers (consisting of covalently linked GP1 and GP2) Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Ebola virus sp. / Strain: Bundibugyo virus Ebola virus sp. / Strain: Bundibugyo virus |
| Molecular weight | Theoretical: 160 KDa |
| Recombinant expression | Organism:   Drosophila melanogaster (fruit fly) / Recombinant cell: S2 / Recombinant plasmid: pMT puro Drosophila melanogaster (fruit fly) / Recombinant cell: S2 / Recombinant plasmid: pMT puro |
-Experimental details
-Structure determination
 Processing Processing |  single particle reconstruction single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris, 150 mM NaCl |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Date | Mar 5, 2015 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 48 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 5512 |
|---|
 Movie
Movie Controller
Controller