[English] 日本語
 Yorodumi
Yorodumi- EMDB-5856: Negative stain reconstruction of HIV envelope glycoprotein BG505 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5856 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
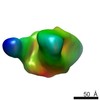
| Title | Negative stain reconstruction of HIV envelope glycoprotein BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab | |||||||||
 Map data Map data | Negative stain reconstruction of HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV / envelope glycoprotein trimer / CAP256 / VRC.26 / V1/V2 directed antibody HIV / envelope glycoprotein trimer / CAP256 / VRC.26 / V1/V2 directed antibody | |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 28.0 Å negative staining / Resolution: 28.0 Å | |||||||||
 Authors Authors | Ward AB / Kim HJ | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Authors: Nicole A Doria-Rose / Chaim A Schramm / Jason Gorman / Penny L Moore / Jinal N Bhiman / Brandon J DeKosky / Michael J Ernandes / Ivelin S Georgiev / Helen J Kim / Marie Pancera / Ryan P ...Authors: Nicole A Doria-Rose / Chaim A Schramm / Jason Gorman / Penny L Moore / Jinal N Bhiman / Brandon J DeKosky / Michael J Ernandes / Ivelin S Georgiev / Helen J Kim / Marie Pancera / Ryan P Staupe / Han R Altae-Tran / Robert T Bailer / Ema T Crooks / Albert Cupo / Aliaksandr Druz / Nigel J Garrett / Kam H Hoi / Rui Kong / Mark K Louder / Nancy S Longo / Krisha McKee / Molati Nonyane / Sijy O'Dell / Ryan S Roark / Rebecca S Rudicell / Stephen D Schmidt / Daniel J Sheward / Cinque Soto / Constantinos Kurt Wibmer / Yongping Yang / Zhenhai Zhang / / James C Mullikin / James M Binley / Rogier W Sanders / Ian A Wilson / John P Moore / Andrew B Ward / George Georgiou / Carolyn Williamson / Salim S Abdool Karim / Lynn Morris / Peter D Kwong / Lawrence Shapiro / John R Mascola /    Abstract: Antibodies capable of neutralizing HIV-1 often target variable regions 1 and 2 (V1V2) of the HIV-1 envelope, but the mechanism of their elicitation has been unclear. Here we define the developmental ...Antibodies capable of neutralizing HIV-1 often target variable regions 1 and 2 (V1V2) of the HIV-1 envelope, but the mechanism of their elicitation has been unclear. Here we define the developmental pathway by which such antibodies are generated and acquire the requisite molecular characteristics for neutralization. Twelve somatically related neutralizing antibodies (CAP256-VRC26.01-12) were isolated from donor CAP256 (from the Centre for the AIDS Programme of Research in South Africa (CAPRISA)); each antibody contained the protruding tyrosine-sulphated, anionic antigen-binding loop (complementarity-determining region (CDR) H3) characteristic of this category of antibodies. Their unmutated ancestor emerged between weeks 30-38 post-infection with a 35-residue CDR H3, and neutralized the virus that superinfected this individual 15 weeks after initial infection. Improved neutralization breadth and potency occurred by week 59 with modest affinity maturation, and was preceded by extensive diversification of the virus population. HIV-1 V1V2-directed neutralizing antibodies can thus develop relatively rapidly through initial selection of B cells with a long CDR H3, and limited subsequent somatic hypermutation. These data provide important insights relevant to HIV-1 vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5856.map.gz emd_5856.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5856-v30.xml emd-5856-v30.xml emd-5856.xml emd-5856.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5856.gif 400_5856.gif 80_5856.gif 80_5856.gif | 22.6 KB 2.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5856 http://ftp.pdbj.org/pub/emdb/structures/EMD-5856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5856 | HTTPS FTP |
-Related structure data
| Related structure data |  4ocrC  4ocsC  4ocwC  4od1C  4od3C  4odhC  4orgC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5856.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5856.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain reconstruction of HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab
| Entire | Name: HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab |
|---|---|
| Components |
|
-Supramolecule #1000: HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab
| Supramolecule | Name: HIV-1 BG505 SOSIP.664 in complex with CAP256-VRC26.09 Fab type: sample / ID: 1000 / Details: Sample was monodisperse / Oligomeric state: One HIV trimer binds one Fab / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: envelope glycoprotein BG505 SOSIP.664
| Macromolecule | Name: envelope glycoprotein BG505 SOSIP.664 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: Trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / synonym: HIV-1 / Location in cell: Membrane Human immunodeficiency virus 1 / synonym: HIV-1 / Location in cell: Membrane |
| Molecular weight | Theoretical: 360 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293S / Recombinant plasmid: pPI4 Homo sapiens (human) / Recombinant cell: HEK 293S / Recombinant plasmid: pPI4 |
-Macromolecule #2: CAP256-VRC26.09 Fab
| Macromolecule | Name: CAP256-VRC26.09 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: heterodimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human / Cell: B cells Homo sapiens (human) / synonym: Human / Cell: B cells |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: IgG1, kappa, lambda Homo sapiens (human) / Recombinant cell: HEK 293F / Recombinant plasmid: IgG1, kappa, lambda |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Grids were adsorbed with protein, blotted, and stained with 2% uranyl formate for 20 seconds. |
| Grid | Details: 400 Cu mesh grid with thin carbon support, glow discharged in natural atmosphere |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Specialist optics | Energy filter - Name: FEI |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 50 |
| Temperature | Min: 292 K / Max: 294 K / Average: 293 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification |
| Date | Oct 18, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 174 / Average electron dose: 25 e/Å2 |
| Tilt angle min | 0 |
- Image processing
Image processing
| Final two d classification | Number classes: 64 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 28.0 Å / Resolution method: OTHER / Software - Name: EMAN, EMAN2, Xmipp, IMAGIC / Details: Map was calculated from a single dataset. / Number images used: 6763 |
| Details | The particles were picked using the automated DoG Picker and put into a stack using the Appion software package. Initial reference free 2D class averages were calculated using particles binned by 2 via Xmipp Clustering 2D Alignment and sorted into classes. A template stack of 44 class averages was used to generate an ab initio model, which was refined using EMAN. |
 Movie
Movie Controller
Controller