+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5564 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
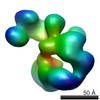
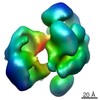
| Title | Compact conformation of human crm1 | |||||||||
 Map data Map data | Compact human crm1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | compact conformation /  crm1 / crm1 /  xpo1 xpo1 | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 20.0 Å negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Doelker N / Blanchet CE / Haselbach D / Voss B / Kappel C / Monecke T / Svergun D / Stark H / Ficner R / Zacharie U ...Doelker N / Blanchet CE / Haselbach D / Voss B / Kappel C / Monecke T / Svergun D / Stark H / Ficner R / Zacharie U / Grubmueller H / Dickmanns A | |||||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Structural determinants and mechanism of mammalian CRM1 allostery. Authors: Nicole Dölker / Clement E Blanchet / Béla Voß / David Haselbach / Christian Kappel / Thomas Monecke / Dmitri I Svergun / Holger Stark / Ralf Ficner / Ulrich Zachariae / Helmut Grubmüller / Achim Dickmanns /  Abstract: Proteins carrying nuclear export signals cooperatively assemble with the export factor CRM1 and the effector protein RanGTP. In lower eukaryotes, this cooperativity is coupled to CRM1 conformational ...Proteins carrying nuclear export signals cooperatively assemble with the export factor CRM1 and the effector protein RanGTP. In lower eukaryotes, this cooperativity is coupled to CRM1 conformational changes; however, it is unknown if mammalian CRM1 maintains its compact conformation or shows similar structural flexibility. Here, combinations of small-angle X-ray solution scattering and electron microscopy experiments with molecular dynamics simulations reveal pronounced conformational flexibility in mammalian CRM1 and demonstrate that RanGTP binding induces association of its N- and C-terminal regions to form a toroid structure. The CRM1 toroid is stabilized mainly by local interactions between the terminal regions, rather than by global strain. The CRM1 acidic loop is key in transmitting the effect of this RanGTP-induced global conformational change to the NES-binding cleft by shifting its population to the open state, which displays enhanced cargo affinity. Cooperative CRM1 export complex assembly thus constitutes a highly dynamic process, encompassing an intricate interplay of global and local structural changes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5564.map.gz emd_5564.map.gz | 568.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5564-v30.xml emd-5564-v30.xml emd-5564.xml emd-5564.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5564_1.jpg emd_5564_1.jpg | 94 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5564 http://ftp.pdbj.org/pub/emdb/structures/EMD-5564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5564 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5564.map.gz / Format: CCP4 / Size: 602.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5564.map.gz / Format: CCP4 / Size: 602.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Compact human crm1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Compact conformation of human crm1
| Entire | Name: Compact conformation of human crm1 |
|---|---|
| Components |
|
-Supramolecule #1000: Compact conformation of human crm1
| Supramolecule | Name: Compact conformation of human crm1 / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: xpo-1
| Macromolecule | Name: xpo-1 / type: protein_or_peptide / ID: 1 / Name.synonym: crm1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 110 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM NaCl, 50 mM HEPES/NaOH, pH 7.5, 2 mM MgOAc, 2 mM DTT |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein were floated on 2% w/v uranyl acetate for 20 seconds. |
| Grid | Details: 200 mesh copper grid with thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Electron beam | Acceleration voltage: 160 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Date | Sep 28, 2012 |
| Image recording | Number real images: 250 |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: imagic / Number images used: 22854 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller