[English] 日本語
 Yorodumi
Yorodumi- EMDB-5381: Cdc6-induced Conformational Changes in ORC Bound To Origin DNA Re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5381 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
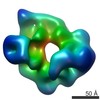
| Title | Cdc6-induced Conformational Changes in ORC Bound To Origin DNA Revealed by Cryo-Electron Microscopy. This map may be mirrored (based on comparison to EMD-5625). | |||||||||
 Map data Map data | cryoEM structure of Yeast Origin Recognition complex with dsDNA and Cdc6. This map may be mirrored (based on comparison to EMD-5625). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ORC /  Cdc6 Cdc6 | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 15.0 Å cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Sun J / Kawakami H / Zech J / Speck C / Stillman B / Li H | |||||||||
 Citation Citation |  Journal: Structure / Year: 2012 Journal: Structure / Year: 2012Title: Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Authors: Jingchuan Sun / Hironori Kawakami / Juergen Zech / Christian Speck / Bruce Stillman / Huilin Li /  Abstract: The eukaryotic origin recognition complex (ORC) interacts with and remodels origins of DNA replication prior to initiation in S phase. Here, we report a single-particle cryo-EM-derived structure of ...The eukaryotic origin recognition complex (ORC) interacts with and remodels origins of DNA replication prior to initiation in S phase. Here, we report a single-particle cryo-EM-derived structure of the supramolecular assembly comprising Saccharomyces cerevisiae ORC, the replication initiation factor Cdc6, and double-stranded ARS1 origin DNA in the presence of ATPγS. The six subunits of ORC are arranged as Orc1:Orc4:Orc5:Orc2:Orc3, with Orc6 binding to Orc2. Cdc6 binding changes the conformation of ORC, in particular reorienting the Orc1 N-terminal BAH domain. Segmentation of the 3D map of ORC-Cdc6 on DNA and docking with the crystal structure of the homologous archaeal Orc1/Cdc6 protein suggest an origin DNA binding model in which the DNA tracks along the interior surface of the crescent-like ORC. Thus, ORC bends and wraps the DNA. This model is consistent with the observation that binding of a single Cdc6 extends the ORC footprint on origin DNA from both ends. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5381.map.gz emd_5381.map.gz | 390.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5381-v30.xml emd-5381-v30.xml emd-5381.xml emd-5381.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5381_1.jpg emd_5381_1.jpg | 27.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5381 http://ftp.pdbj.org/pub/emdb/structures/EMD-5381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5381 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5381.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5381.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM structure of Yeast Origin Recognition complex with dsDNA and Cdc6. This map may be mirrored (based on comparison to EMD-5625). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.23 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yeast Origin Recognition Complex with dsDNA and Cdc6
| Entire | Name: Yeast Origin Recognition Complex with dsDNA and Cdc6 |
|---|---|
| Components |
|
-Supramolecule #1000: Yeast Origin Recognition Complex with dsDNA and Cdc6
| Supramolecule | Name: Yeast Origin Recognition Complex with dsDNA and Cdc6 / type: sample / ID: 1000 Details: ORC and Cdc6 are mixed with 66 bp DNA with ARS1 in ATPrS Number unique components: 8 |
|---|---|
| Molecular weight | Experimental: 500 KDa / Theoretical: 500 KDa |
-Macromolecule #1: Yeast Origin Recognition Complex and Cdc6
| Macromolecule | Name: Yeast Origin Recognition Complex and Cdc6 / type: protein_or_peptide / ID: 1 / Name.synonym: ORC, Cdc6 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / synonym: ORC, Cdc6 Saccharomyces cerevisiae (brewer's yeast) / synonym: ORC, Cdc6 |
| Molecular weight | Experimental: 500 KDa / Theoretical: 500 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Recombinant plasmid: pCITE-2a Saccharomyces cerevisiae (brewer's yeast) / Recombinant plasmid: pCITE-2a |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM HEPES/KOH, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 100 mM KGlu |
| Grid | Details: 300 mesh lacey grid with thin continuous carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Instrument: OTHER / Details: Vitrification instrument: FEI Vitrobot / Method: Blot 5 seconds |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 60000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Gatan 626200 / Specimen holder model: GATAN LIQUID NITROGEN |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON COOLSCAN / Digitization - Sampling interval: 6.35 µm / Average electron dose: 15 e/Å2 / Bits/pixel: 8 |
- Image processing
Image processing
| CTF correction | Details: Each particle set |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 15.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN1.8 / Number images used: 54000 |
 Movie
Movie Controller
Controller