[English] 日本語
 Yorodumi
Yorodumi- EMDB-5325: Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5325 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
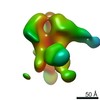
| Title | Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with 17b | |||||||||
 Map data Map data | Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with 17b | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV/AIDS vaccine / HIV/AIDS vaccine /  immunogen / HIV glycoprotein structure / gp140 trimer / immunogen / HIV glycoprotein structure / gp140 trimer /  cryo-electron tomography / sub-tomogram averaging / cryo-electron tomography / sub-tomogram averaging /  cryo-electron microscopy cryo-electron microscopy | |||||||||
| Biological species |    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 20.0 Å cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Harris A / Borgnia MJ / Shi D / Bartesaghi A / He H / Pejchal R / Kang YK / Depetris R / Marozsan AJ / Sanders RW ...Harris A / Borgnia MJ / Shi D / Bartesaghi A / He H / Pejchal R / Kang YK / Depetris R / Marozsan AJ / Sanders RW / Klasse PJ / Milne JL / Wilson IA / Olson WC / Moore JP / Subramaniam S | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Authors: Audray Harris / Mario J Borgnia / Dan Shi / Alberto Bartesaghi / Haifeng He / Robert Pejchal / Yun Kenneth Kang / Rafael Depetris / Andre J Marozsan / Rogier W Sanders / Per Johan Klasse / ...Authors: Audray Harris / Mario J Borgnia / Dan Shi / Alberto Bartesaghi / Haifeng He / Robert Pejchal / Yun Kenneth Kang / Rafael Depetris / Andre J Marozsan / Rogier W Sanders / Per Johan Klasse / Jacqueline L S Milne / Ian A Wilson / William C Olson / John P Moore / Sriram Subramaniam /  Abstract: The initial step in HIV-1 infection occurs with the binding of cell surface CD4 to trimeric HIV-1 envelope glycoproteins (Env), a heterodimer of a transmembrane glycoprotein (gp41) and a surface ...The initial step in HIV-1 infection occurs with the binding of cell surface CD4 to trimeric HIV-1 envelope glycoproteins (Env), a heterodimer of a transmembrane glycoprotein (gp41) and a surface glycoprotein (gp120). The design of soluble versions of trimeric Env that display structural and functional properties similar to those observed on intact viruses is highly desirable from the viewpoint of designing immunogens that could be effective as vaccines against HIV/AIDS. Using cryoelectron tomography combined with subvolume averaging, we have analyzed the structure of SOSIP gp140 trimers, which are cleaved, solubilized versions of the ectodomain of trimeric HIV-1 Env. We show that unliganded gp140 trimers adopt a quaternary arrangement similar to that displayed by native unliganded trimers on the surface of intact HIV-1 virions. When complexed with soluble CD4, Fab 17b, which binds to gp120 at its chemokine coreceptor binding site, or both soluble CD4 and 17b Fab, gp140 trimers display an open conformation in which there is an outward rotation and displacement of each gp120 protomer. We demonstrate that the molecular arrangements of gp120 trimers in the closed and open conformations of the soluble trimer are the same as those observed for the closed and open states, respectively, of trimeric gp120 on intact HIV-1 BaL virions, establishing that soluble gp140 trimers can be designed to mimic the quaternary structural transitions displayed by native trimeric Env. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5325.map.gz emd_5325.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5325-v30.xml emd-5325-v30.xml emd-5325.xml emd-5325.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5325_1.jpg emd_5325_1.jpg | 29.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5325 http://ftp.pdbj.org/pub/emdb/structures/EMD-5325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5325 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5325 | HTTPS FTP |
-Related structure data
| Related structure data |  1930C  5320C  5321C  5322C  5323C  5324C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5325.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5325.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with 17b | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SOSIP gp140 KNH1144 17b
| Entire | Name: SOSIP gp140 KNH1144 17b |
|---|---|
| Components |
|
-Supramolecule #1000: SOSIP gp140 KNH1144 17b
| Supramolecule | Name: SOSIP gp140 KNH1144 17b / type: sample / ID: 1000 / Oligomeric state: trimeric / Number unique components: 4 |
|---|
-Macromolecule #1: SOSIP gp140 KNH1144 17b
| Macromolecule | Name: SOSIP gp140 KNH1144 17b / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller