[English] 日本語
 Yorodumi
Yorodumi- EMDB-5288: The single particle reconstruction of the human Toll-like recepto... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5288 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
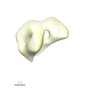
| Title | The single particle reconstruction of the human Toll-like receptor 5 ectodomain in detergent | |||||||||
 Map data Map data | This is the volume of the hTLR5 ectodomain. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Toll-like receptor 5 / Toll-like receptor 5 /  ectodomain ectodomain | |||||||||
| Function / homology |  Toll-like receptor Toll-like receptor Function and homology information Function and homology information | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 26.0 Å negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Zhou K / Kanai R / Lee P / Wang HW / Modis Y | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2012 Journal: J Struct Biol / Year: 2012Title: Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin. Authors: Kaifeng Zhou / Ryuta Kanai / Phong Lee / Hong-Wei Wang / Yorgo Modis /  Abstract: The structure of full-length human TLR5 determined by electron microscopy single-particle image reconstruction at 26Å resolution shows that TLR5 forms an asymmetric homodimer via ectodomain ...The structure of full-length human TLR5 determined by electron microscopy single-particle image reconstruction at 26Å resolution shows that TLR5 forms an asymmetric homodimer via ectodomain interactions. The structure shows that like TLR9, TLR5 dimerizes in the absence of ligand. The asymmetry of the dimer suggests that TLR5 may recognize two flagellin molecules cooperatively to establish an optimal flagellin response threshold. A TLR5 homology model was generated and fitted into the electron microscopy structure. All seven predicted N-linked glycosylation sites are exposed on the molecular surface, away from the dimer interface. Glycosylation at the first five sites was confirmed by tandem mass spectrometry. Two aspartate residues proposed to interact with flagellin (Asp294 and Asp366) are sterically occluded by a glycan at position 342. In contrast, the central region of the ectodomains near the dimer interface is unobstructed by glycans. Ligand binding in this region would be consistent with the ligand binding sites of other TLRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5288.map.gz emd_5288.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5288-v30.xml emd-5288-v30.xml emd-5288.xml emd-5288.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5288_1.png emd_5288_1.png | 300.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5288 http://ftp.pdbj.org/pub/emdb/structures/EMD-5288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5288 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5288 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5288.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5288.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the volume of the hTLR5 ectodomain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.14 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : toll-like receptor 5 ectodomain
| Entire | Name: toll-like receptor 5 ectodomain |
|---|---|
| Components |
|
-Supramolecule #1000: toll-like receptor 5 ectodomain
| Supramolecule | Name: toll-like receptor 5 ectodomain / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: dimer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 200 KDa / Theoretical: 200 KDa / Method: gel filtration |
-Macromolecule #1: Toll-like receptor
| Macromolecule | Name: Toll-like receptor / type: protein_or_peptide / ID: 1 / Name.synonym: TLR5 / Number of copies: 2 / Oligomeric state: dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human / Cell: Sf9 insect cells / Organelle: membrane / Location in cell: plasma membrane Homo sapiens (human) / synonym: Human / Cell: Sf9 insect cells / Organelle: membrane / Location in cell: plasma membrane |
| Molecular weight | Experimental: 200 KDa / Theoretical: 200 KDa |
| Recombinant expression | Organism: Sf9 insect cells / Recombinant plasmid: pAcGP67-A |
| Sequence | InterPro:  Toll-like receptor Toll-like receptor |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.005 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 10 mM TEA, 0.15 M NaCl |
| Staining | Type: NEGATIVE Details: The protein solution was applied onto a thin-carbon-covered holey carbon copper grid and negatively stained in 1% uranyl-formate solution for 1 minute. |
| Grid | Details: thin carbon covered home-made holey carbon on 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 52000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Date | Aug 1, 2010 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 50 |
- Image processing
Image processing
| Final two d classification | Number classes: 100 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC SPIDER Details: Final maps were calculated from all the particles by back-projection reconstruction. Number images used: 3087 |
 Movie
Movie Controller
Controller