[English] 日本語
 Yorodumi
Yorodumi- EMDB-5272: Molecular Structure of Unliganded Native SIVmneE11S gp120 trimer:... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5272 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
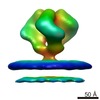
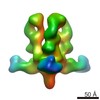
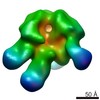
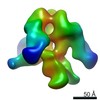
| Title | Molecular Structure of Unliganded Native SIVmneE11S gp120 trimer: Spike region | |||||||||
 Map data Map data | 3D Average of Envelope Glycoproteins from SIVmneE11S - structures determined by cryo-electron tomography combined with 3D averaging | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Antigens / CD4 / Antigens / CD4 /  cryo-electron microscopy / cryo-electron microscopy /  HIV / SIV / envelope glycoprotein / HIV / SIV / envelope glycoprotein /  gp120 / gp120 /  gp41 / gp41 /  HIV-1 / HIV-1 /  Immunoglobulin Fab Fragments Immunoglobulin Fab Fragments | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 20.0 Å cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | White TA / Bartesaghi A / Borgnia M / Subramaniam S | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2010 Journal: PLoS Pathog / Year: 2010Title: Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. Authors: Tommi A White / Alberto Bartesaghi / Mario J Borgnia / Joel R Meyerson / M Jason V de la Cruz / Julian W Bess / Rachna Nandwani / James A Hoxie / Jeffrey D Lifson / Jacqueline L S Milne / Sriram Subramaniam /  Abstract: The initial step in target cell infection by human, and the closely related simian immunodeficiency viruses (HIV and SIV, respectively) occurs with the binding of trimeric envelope glycoproteins (Env) ...The initial step in target cell infection by human, and the closely related simian immunodeficiency viruses (HIV and SIV, respectively) occurs with the binding of trimeric envelope glycoproteins (Env), composed of heterodimers of the viral transmembrane glycoprotein (gp41) and surface glycoprotein (gp120) to target T-cells. Knowledge of the molecular structure of trimeric Env on intact viruses is important both for understanding the molecular mechanisms underlying virus-cell interactions and for the design of effective immunogen-based vaccines to combat HIV/AIDS. Previous analyses of intact HIV-1 BaL virions have already resulted in structures of trimeric Env in unliganded and CD4-liganded states at ~20 Å resolution. Here, we show that the molecular architectures of trimeric Env from SIVmneE11S, SIVmac239 and HIV-1 R3A strains are closely comparable to that previously determined for HIV-1 BaL, with the V1 and V2 variable loops located at the apex of the spike, close to the contact zone between virus and cell. The location of the V1/V2 loops in trimeric Env was definitively confirmed by structural analysis of HIV-1 R3A virions engineered to express Env with deletion of these loops. Strikingly, in SIV CP-MAC, a CD4-independent strain, trimeric Env is in a constitutively "open" conformation with gp120 trimers splayed out in a conformation similar to that seen for HIV-1 BaL Env when it is complexed with sCD4 and the CD4i antibody 17b. Our findings suggest a structural explanation for the molecular mechanism of CD4-independent viral entry and further establish that cryo-electron tomography can be used to discover distinct, functionally relevant quaternary structures of Env displayed on intact viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5272.map.gz emd_5272.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5272-v30.xml emd-5272-v30.xml emd-5272.xml emd-5272.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5272_1.jpg emd_5272_1.jpg | 77.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5272 http://ftp.pdbj.org/pub/emdb/structures/EMD-5272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5272 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5272.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5272.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D Average of Envelope Glycoproteins from SIVmneE11S - structures determined by cryo-electron tomography combined with 3D averaging | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SIVmneE11S virus
| Entire | Name: SIVmneE11S virus |
|---|---|
| Components |
|
-Supramolecule #1000: SIVmneE11S virus
| Supramolecule | Name: SIVmneE11S virus / type: sample / ID: 1000 Details: Structures determined by cryo-electron tomography combined with 3D averaging Oligomeric state: trimer / Number unique components: 1 |
|---|
-Macromolecule #1: envelope glycoprotein from Simian Immunodeficiency virus (clone E...
| Macromolecule | Name: envelope glycoprotein from Simian Immunodeficiency virus (clone E11S) infecting Macaca nemestrina type: protein_or_peptide / ID: 1 / Name.synonym: Env Details: Envelope glycoproteins present on the surface of intact virions. Virus infecting Macaca nemestrina (Pig-tailed macaque) Oligomeric state: trimer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) / Strain: SIVmneE11S / synonym: Simian Immunodeficiency Virus / Cell: HUT-78 / Location in cell: viral membrane |
| Molecular weight | Theoretical: 480 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: TNE Buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA) |
|---|---|
| Grid | Details: 200 mesh Quantifoil Multi A |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 77 K / Instrument: FEI VITROBOT MARK III Details: Vitrification instrument: Mark III Vitrobot (FEI, Netherlands) Method: blot for 6 seconds, at 25 C, 100 percent humidity, blot offset of -2, into an ethane slurry cooled by liquid nitrogen |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 34000 Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 34000 |
| Specialist optics | Energy filter - Name: GATAN GIF / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder: Cartridge / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -70 ° / Tilt series - Axis1 - Max angle: 70 ° |
| Temperature | Min: 81 K / Max: 82 K / Average: 81 K |
| Details | Areas were selected for proper ice thickness (about 200 nm). |
| Date | May 14, 2008 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Average electron dose: 150 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name:  IMOD IMODDetails: Please see A. Bartesaghi, et al. Journal of Structural Biology, 2008 |
|---|---|
| Details | Average tomographic tilt angle increment: 1. |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  UCSF Chimera UCSF Chimera |
| Details | Protocol: Rigid body. Automated fitting procedures |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
 Movie
Movie Controller
Controller