+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5224 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human 80S ribosome in situ, untreated Eukaryotic ribosome Eukaryotic ribosome | |||||||||
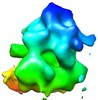
 Map data Map data | This is a map of a tomographic average of a human 80S ribosome in situ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  human 80S ribosome / human 80S ribosome /  puromycin / puromycin /  in situ / in situ /  cytosol / cytosol /  polysome / polysome /  polyribosome / polyribosome /  protein synthesis / protein synthesis /  translation / 3D cryoEM / translation / 3D cryoEM /  tomography / cellular tomography tomography / cellular tomography | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | subtomogram averaging /  cryo EM / cryo EM /  negative staining / Resolution: 39.0 Å negative staining / Resolution: 39.0 Å | |||||||||
 Authors Authors | Brandt F / Carlson L-A / Hartl FU / Baumeister W / Grunewald K | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2010 Journal: Mol Cell / Year: 2010Title: The three-dimensional organization of polyribosomes in intact human cells. Authors: Florian Brandt / Lars-Anders Carlson / F Ulrich Hartl / Wolfgang Baumeister / Kay Grünewald /  Abstract: Structural studies have provided detailed insights into different functional states of the ribosome and its interaction with factors involved in nascent peptide folding, processing, and targeting. ...Structural studies have provided detailed insights into different functional states of the ribosome and its interaction with factors involved in nascent peptide folding, processing, and targeting. However, how the translational machinery is organized spatially in native cellular environments is not yet well understood. Here we have mapped individual ribosomes in electron tomograms of intact human cells by template matching and determined the average structure of the ribosome in situ. Characteristic features of active ribosomes in the cellular environment were assigned to the tRNA channel, elongation factors, and additional densities near the peptide tunnel. Importantly, the relative spatial configuration of neighboring ribosomes in the cell is clearly nonrandom. The preferred configurations are specific for active polysomes and were largely abrogated in puromycin-treated control cells. The distinct neighbor orientations found in situ resemble configurations of bacterial polysomes in vitro, indicating a conserved supramolecular organization with implications for nascent polypeptide folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5224.map.gz emd_5224.map.gz | 397.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5224-v30.xml emd-5224-v30.xml emd-5224.xml emd-5224.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5224_1.jpg emd_5224_1.jpg | 58.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5224 http://ftp.pdbj.org/pub/emdb/structures/EMD-5224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5224 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5224 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5224.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5224.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of a tomographic average of a human 80S ribosome in situ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.21 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human 80S ribosome in situ, untreated
| Entire | Name: Human 80S ribosome in situ, untreated Eukaryotic ribosome Eukaryotic ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: Human 80S ribosome in situ, untreated
| Supramolecule | Name: Human 80S ribosome in situ, untreated / type: sample / ID: 1000 / Details: Cytosolic ribosomes in situ Oligomeric state: One 80S ribosome within mixed cellular polysomes Number unique components: 1 |
|---|---|
| Molecular weight | Method: Sedimentation, 80S |
-Supramolecule #1: cytosolic 80S ribosome
| Supramolecule | Name: cytosolic 80S ribosome / type: complex / ID: 1 / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: human Homo sapiens (human) / synonym: human |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: Cellular medium, DMEM (Gibco) supplemented with 10% foetal calf serum, 37C and 5% CO2 |
|---|---|
| Staining | Type: NEGATIVE / Details: Cells grown on grids, vitrification |
| Grid | Details: C-flat 2/1, holey carbon gold grid |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 77 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: plunger. Vitrification carried out in air Method: Blot for 2 s before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 17500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 17500 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 17500 |
| Specialist optics | Energy filter - Name: GIF2002 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder. Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -65 ° / Tilt series - Axis1 - Max angle: 65 ° |
| Temperature | Min: 77 K / Average: 77 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 50,000 times magnification |
| Date | Jul 7, 2008 |
| Image recording | Category: CCD / Film or detector model: GATAN MULTISCAN / Average electron dose: 80 e/Å2 |
- Image processing
Image processing
| Final 3D classification | Number classes: 1 |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 39.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EM / Details: exact weighting / Number subtomograms used: 1911 |
| Details | Individual particle subvolumes were automatically selected by CCC threshold. |
 Movie
Movie Controller
Controller