[English] 日本語
 Yorodumi
Yorodumi- EMDB-5154: Negative stain EM of the recombinant Acidianus tengchongensis cha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5154 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
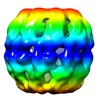
| Title | Negative stain EM of the recombinant Acidianus tengchongensis chaperonin beta. | |||||||||
 Map data Map data | This is the negative stain microscopy reconstruction of the recombinant thermosome beta from Acidianus tengchongensis | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rATcpn-beta / Group II chaperonin /  Thermosome Thermosome | |||||||||
| Biological species |   Acidianus tengchongensis (archaea) Acidianus tengchongensis (archaea) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 14.0 Å negative staining / Resolution: 14.0 Å | |||||||||
 Authors Authors | Huo Y / Hu Z / Zhang K / Wang L / Zhai Y / Zhou Q / Lander G / Zhu J / He Y / Pang X ...Huo Y / Hu Z / Zhang K / Wang L / Zhai Y / Zhou Q / Lander G / Zhu J / He Y / Pang X / Xu W / Bartlam M / Dong Z / Sun F | |||||||||
 Citation Citation |  Journal: Structure / Year: 2010 Journal: Structure / Year: 2010Title: Crystal structure of group II chaperonin in the open state. Authors: Yanwu Huo / Zhongjun Hu / Kai Zhang / Li Wang / Yujia Zhai / Qiangjun Zhou / Gabe Lander / Jiang Zhu / Yongzhi He / Xiaoyun Pang / Wei Xu / Mark Bartlam / Zhiyang Dong / Fei Sun /  Abstract: Thermosomes are group II chaperonins responsible for protein refolding in an ATP-dependent manner. Little is known regarding the conformational changes of thermosomes during their functional cycle ...Thermosomes are group II chaperonins responsible for protein refolding in an ATP-dependent manner. Little is known regarding the conformational changes of thermosomes during their functional cycle due to a lack of high-resolution structure in the open state. Here, we report the first complete crystal structure of thermosome (rATcpnβ) in the open state from Acidianus tengchongensis. There is a ∼30° rotation of the apical and lid domains compared with the previous closed structure. Besides, the structure reveals a conspicuous hydrophobic patch in the lid domain, and residues locating in this patch are conserved across species. Both the closed and open forms of rATcpnβ were also reconstructed by electron microscopy (EM). Structural fitting revealed the detailed conformational change from the open to the closed state. Structural comparison as well as protease K digestion indicated only ATP binding without hydrolysis does not induce chamber closure of thermosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5154.map.gz emd_5154.map.gz | 13.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5154-v30.xml emd-5154-v30.xml emd-5154.xml emd-5154.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5154_1.png emd_5154_1.png | 186.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5154 http://ftp.pdbj.org/pub/emdb/structures/EMD-5154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5154 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5154.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5154.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the negative stain microscopy reconstruction of the recombinant thermosome beta from Acidianus tengchongensis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative stain EM of the recombinant Acidianus tengchongensis cha...
| Entire | Name: Negative stain EM of the recombinant Acidianus tengchongensis chaperonin beta |
|---|---|
| Components |
|
-Supramolecule #1000: Negative stain EM of the recombinant Acidianus tengchongensis cha...
| Supramolecule | Name: Negative stain EM of the recombinant Acidianus tengchongensis chaperonin beta type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: octadecamer / Number unique components: 18 |
|---|---|
| Molecular weight | Experimental: 1.08 MDa / Theoretical: 1.08 MDa |
-Macromolecule #1: rATcpn-beta
| Macromolecule | Name: rATcpn-beta / type: protein_or_peptide / ID: 1 / Name.synonym: chaperonin beta / Number of copies: 18 / Oligomeric state: 18mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Acidianus tengchongensis (archaea) / Strain: S5 / Location in cell: cytoplasm Acidianus tengchongensis (archaea) / Strain: S5 / Location in cell: cytoplasm |
| Molecular weight | Theoretical: 1.08 MDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant plasmid: pET23b Escherichia coli (E. coli) / Recombinant plasmid: pET23b |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 4% w/v uranyl acetate for 25 seconds. |
| Grid | Details: 200 mesh gold grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 70000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 70000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Digitization - Sampling interval: 14 µm / Number real images: 100 / Bits/pixel: 16 |
- Image processing
Image processing
| CTF correction | Details: each photo |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.0 Å / Resolution method: OTHER / Software - Name: EMAN / Number images used: 2328 |
| Details | The particles were selected by hand. |
 Movie
Movie Controller
Controller