[English] 日本語
 Yorodumi
Yorodumi- EMDB-2928: Negative stain structure of a type 6 secretion system membrane co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2928 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
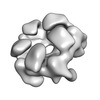
| Title | Negative stain structure of a type 6 secretion system membrane core complex | |||||||||
 Map data Map data | Structure of a T6SS membrane core complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial secretion /  virulence virulence | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Escherichia coli (strain 55989 / EAEC) (bacteria) Escherichia coli (strain 55989 / EAEC) (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 11.5 Å negative staining / Resolution: 11.5 Å | |||||||||
 Authors Authors | Durand E / Fronzes R | |||||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Biogenesis and structure of a type VI secretion membrane core complex. Authors: Eric Durand / Van Son Nguyen / Abdelrahim Zoued / Laureen Logger / Gérard Péhau-Arnaudet / Marie-Stéphanie Aschtgen / Silvia Spinelli / Aline Desmyter / Benjamin Bardiaux / Annick ...Authors: Eric Durand / Van Son Nguyen / Abdelrahim Zoued / Laureen Logger / Gérard Péhau-Arnaudet / Marie-Stéphanie Aschtgen / Silvia Spinelli / Aline Desmyter / Benjamin Bardiaux / Annick Dujeancourt / Alain Roussel / Christian Cambillau / Eric Cascales / Rémi Fronzes /  Abstract: Bacteria share their ecological niches with other microbes. The bacterial type VI secretion system is one of the key players in microbial competition, as well as being an important virulence ...Bacteria share their ecological niches with other microbes. The bacterial type VI secretion system is one of the key players in microbial competition, as well as being an important virulence determinant during bacterial infections. It assembles a nano-crossbow-like structure in the cytoplasm of the attacker cell that propels an arrow made of a haemolysin co-regulated protein (Hcp) tube and a valine-glycine repeat protein G (VgrG) spike and punctures the prey's cell wall. The nano-crossbow is stably anchored to the cell envelope of the attacker by a membrane core complex. Here we show that this complex is assembled by the sequential addition of three type VI subunits (Tss)-TssJ, TssM and TssL-and present a structure of the fully assembled complex at 11.6 Å resolution, determined by negative-stain electron microscopy. With overall C5 symmetry, this 1.7-megadalton complex comprises a large base in the cytoplasm. It extends in the periplasm via ten arches to form a double-ring structure containing the carboxy-terminal domain of TssM (TssMct) and TssJ that is anchored in the outer membrane. The crystal structure of the TssMct-TssJ complex coupled to whole-cell accessibility studies suggest that large conformational changes induce transient pore formation in the outer membrane, allowing passage of the attacking Hcp tube/VgrG spike. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2928.map.gz emd_2928.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2928-v30.xml emd-2928-v30.xml emd-2928.xml emd-2928.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2928_image.png EMD-2928_image.png | 68 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2928 http://ftp.pdbj.org/pub/emdb/structures/EMD-2928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2928 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2928.map.gz / Format: CCP4 / Size: 81.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2928.map.gz / Format: CCP4 / Size: 81.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of a T6SS membrane core complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative stain structure of the TssJ-TssM-TssL type 6 secretion m...
| Entire | Name: Negative stain structure of the TssJ-TssM-TssL type 6 secretion membrane core complex |
|---|---|
| Components |
|
-Supramolecule #1000: Negative stain structure of the TssJ-TssM-TssL type 6 secretion m...
| Supramolecule | Name: Negative stain structure of the TssJ-TssM-TssL type 6 secretion membrane core complex type: sample / ID: 1000 / Oligomeric state: Decamer / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1.7 MDa / Theoretical: 1.7 MDa Method: Relative stoichometry of the individual components is 1:1:1 for TssJ:TssM:TssL. Docking of the crystal structures of some parts of the complex indicates 10 copies of each protein. |
-Macromolecule #1: TssJ
| Macromolecule | Name: TssJ / type: protein_or_peptide / ID: 1 / Name.synonym: T6SS_SciN / Number of copies: 10 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: outer membrane Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: outer membrane |
| Molecular weight | Experimental: 18.4 KDa / Theoretical: 18.4 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet |
| Sequence | UniProtKB:  Type VI secretion system lipoprotein TssJ / InterPro: Type VI secretion system lipoprotein TssJ / InterPro:  Type VI secretion system, lipoprotein SciN Type VI secretion system, lipoprotein SciN |
-Macromolecule #2: TssM
| Macromolecule | Name: TssM / type: protein_or_peptide / ID: 2 / Name.synonym: T6SS_SciS / Number of copies: 10 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: inner membrane Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: inner membrane |
| Molecular weight | Experimental: 130 KDa / Theoretical: 130 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet |
| Sequence | UniProtKB:  Putative type VI secretion protein Putative type VI secretion proteinInterPro:  Type VI secretion system IcmF, C-terminal, IcmF-related Type VI secretion system IcmF, C-terminal, IcmF-related |
-Macromolecule #3: TssL
| Macromolecule | Name: TssL / type: protein_or_peptide / ID: 3 / Name.synonym: T6SS_SciP / Number of copies: 10 / Oligomeric state: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: inner membrane Escherichia coli (strain 55989 / EAEC) (bacteria) / Strain: strain 55989 / EAEC / synonym: Enteroaggregative Escherichia coli / Location in cell: inner membrane |
| Molecular weight | Experimental: 24 KDa / Theoretical: 24 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet Escherichia coli BL21(DE3) (bacteria) / Recombinant plasmid: pRSF-Duet |
| Sequence | UniProtKB:  Type VI secretion protein / InterPro: Type IV / VI secretion system, DotU Type VI secretion protein / InterPro: Type IV / VI secretion system, DotU |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.025% w/v Decyl Maltose Neopentyl Glycol (DM-NPG) |
| Staining | Type: NEGATIVE Details: Nine microlitres of TssJLM complex sample was applied to glow-discharged carbon-coated copper grids (Agar Scientific). After 30 sec of absorption, the sample was blotted, washed with three ...Details: Nine microlitres of TssJLM complex sample was applied to glow-discharged carbon-coated copper grids (Agar Scientific). After 30 sec of absorption, the sample was blotted, washed with three drops of water and then stained with 2% uranyl acetate. |
| Grid | Details: 400 mesh carbon-coated copper grids |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 68100 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.1 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.1 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: OTHER |
| Alignment procedure | Legacy - Astigmatism: Astigmatism was checked during imaging and during CTF correction of the micrographs. Micrographs is astigmatism >10% were discarded. |
| Date | Nov 20, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 850 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: OTHER / Software - Name: EMAN2, Relion / Number images used: 16738 ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: OTHER / Software - Name: EMAN2, Relion / Number images used: 16738 |
| Details | A total of 72,146 particles were automatically selected from 1,200 independent images and extracted within boxes of 280 pixels x 280 pixels using EMAN2/BOXER. The CTF was estimated and corrected by phase flipping using EMAN2 (e2ctf). All two- and three-dimensional (2D and 3D) .classifications and refinements were performed using RELION 1.3 . We used three rounds of reference-free 2D class averaging to clean up the automatically selected dataset. Only highly populated classes displaying high-resolution features were conserved during this procedure and a final dataset of 26,544 particles was assembled. An initial 3D-model was generated in EMAN2 using using 30 classes. 3D classification was then performed in Relion with 5 classes. The particles corresponding to most populated class (~ 16,738) were used for refinement. Relion auto-refine procedure was used to obtain a final reconstruction at 11.56-A resolution after masking and with C5 symmetry imposed. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion, and FSC curve were corrected for the effects of a soft mask on the FSC curve using high-resolution noise substitution . A subregion of the whole reconstruction was further refined by applying a local mask in Relion 3Drefine. |
 Movie
Movie Controller
Controller