[English] 日本語
 Yorodumi
Yorodumi- EMDB-2551: Electron cryo-microscopy of the PTC3 holotoxin complex (TcdA1, Tc... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2551 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
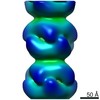
| Title | Electron cryo-microscopy of the PTC3 holotoxin complex (TcdA1, TcdB2, TccC3) in prepore state | |||||||||
 Map data Map data | PTC3 Holotoxin Complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photorabdus / Tc toxin / ABC toxin / cargo-delivery | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 9.0 Å cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Gatsogiannis C / Meusch D / Efremov RG / Lang AE / Hofnagel O / Vetter IR / Aktories K / Raunser S | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Mechanism of Tc toxin action revealed in molecular detail. Authors: Dominic Meusch / Christos Gatsogiannis / Rouslan G Efremov / Alexander E Lang / Oliver Hofnagel / Ingrid R Vetter / Klaus Aktories / Stefan Raunser /  Abstract: Tripartite Tc toxin complexes of bacterial pathogens perforate the host membrane and translocate toxic enzymes into the host cell, including in humans. The underlying mechanism is complex but poorly ...Tripartite Tc toxin complexes of bacterial pathogens perforate the host membrane and translocate toxic enzymes into the host cell, including in humans. The underlying mechanism is complex but poorly understood. Here we report the first, to our knowledge, high-resolution structures of a TcA subunit in its prepore and pore state and of a complete 1.7 megadalton Tc complex. The structures reveal that, in addition to a translocation channel, TcA forms four receptor-binding sites and a neuraminidase-like region, which are important for its host specificity. pH-induced opening of the shell releases an entropic spring that drives the injection of the TcA channel into the membrane. Binding of TcB/TcC to TcA opens a gate formed by a six-bladed β-propeller and results in a continuous protein translocation channel, whose architecture and properties suggest a novel mode of protein unfolding and translocation. Our results allow us to understand key steps of infections involving Tc toxins at the molecular level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2551.map.gz emd_2551.map.gz | 457.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2551-v30.xml emd-2551-v30.xml emd-2551.xml emd-2551.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2551.png emd_2551.png | 976.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2551 http://ftp.pdbj.org/pub/emdb/structures/EMD-2551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2551 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2551.map.gz / Format: CCP4 / Size: 500 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2551.map.gz / Format: CCP4 / Size: 500 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PTC3 Holotoxin Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PTC3 (TcdA1, TcdB2, TccC3)
| Entire | Name: PTC3 (TcdA1, TcdB2, TccC3) |
|---|---|
| Components |
|
-Supramolecule #1000: PTC3 (TcdA1, TcdB2, TccC3)
| Supramolecule | Name: PTC3 (TcdA1, TcdB2, TccC3) / type: sample / ID: 1000 Oligomeric state: TcdB2/TccC3 bind to one homopentamer of TcdA1 Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1.68 MDa / Theoretical: 1.68 MDa |
-Macromolecule #1: TcdA1
| Macromolecule | Name: TcdA1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: Pentamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Experimental: 283 KDa / Theoretical: 283 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | UniProtKB: TcdA1 |
-Macromolecule #2: TccC3
| Macromolecule | Name: TccC3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Theoretical: 109 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | UniProtKB: Insecticidal toxin complex protein TccC3 |
-Macromolecule #3: TcdB2
| Macromolecule | Name: TcdB2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Experimental: 166 KDa / Theoretical: 166 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | UniProtKB: Insecticidal toxin complex protein TcdB2 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 5 Details: 50 mM MES, 100 mM NaCl, 0.05% Tween-20, 5% glycerol |
| Grid | Details: C-Flat 2/1-4C copper 400 mesh, with additional thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 100 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 1 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 124472 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 4.1 mm / Nominal defocus max: 0.0032 µm / Nominal defocus min: 0.0004 µm / Nominal magnification: 60000 Bright-field microscopy / Cs: 4.1 mm / Nominal defocus max: 0.0032 µm / Nominal defocus min: 0.0004 µm / Nominal magnification: 60000 |
| Specialist optics | Energy filter - Name: in-column Omega filter / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 15.0 eV |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER |
| Date | Nov 19, 2012 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F816 (8k x 8k) / Number real images: 1250 / Average electron dose: 20 e/Å2 |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: OTHER / Software - Name: Sparx Details: TcdA1 component of the complex has an average resolution of 7.4 A, whereas the TcdB2/TccC3 components an average resolution of 11 A. Local resolution calculations were performed using a 20 A ...Details: TcdA1 component of the complex has an average resolution of 7.4 A, whereas the TcdB2/TccC3 components an average resolution of 11 A. Local resolution calculations were performed using a 20 A sphere for windowed-FSC calculations at 0.5. Number images used: 43000 |
| Details | Partial-symmetry projection matching approach (after each refinement round, C5 symmetry was applied for the TcdA1 component). |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller