[English] 日本語
 Yorodumi
Yorodumi- EMDB-2216: DOLORS: Versatile Strategy for Internal Labeling and Domain Local... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2216 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DOLORS: Versatile Strategy for Internal Labeling and Domain Localization in Electron Microscopy | |||||||||
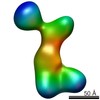
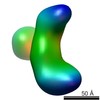
 Map data Map data | RNaseIIIb-labeled Dicer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dicer Enzyme /  Ribonuclease III / MicroRNA Processing / Single Particle Electron Microscopy Ribonuclease III / MicroRNA Processing / Single Particle Electron Microscopy | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining negative staining | |||||||||
 Authors Authors | Lau PW / Potter CS / Carragher B / MacRae IJ | |||||||||
 Citation Citation |  Journal: Structure / Year: 2012 Journal: Structure / Year: 2012Title: DOLORS: versatile strategy for internal labeling and domain localization in electron microscopy. Authors: Pick-Wei Lau / Clinton S Potter / Bridget Carragher / Ian J MacRae /  Abstract: Single-particle electron microscopy (EM) is a powerful tool for studying the structures of large biological molecules. However, the achievable resolution does not always allow for direct recognition ...Single-particle electron microscopy (EM) is a powerful tool for studying the structures of large biological molecules. However, the achievable resolution does not always allow for direct recognition of individual protein domains. Labels that can be visualized by EM have been developed for protein termini, but tagging internal domains remains a challenge. We describe a robust strategy for determining the position of internal sites within EM maps, termed domain localization by RCT sampling (DOLORS). DOLORS uses monovalent streptavidin added posttranslationally to tagged sites in the target protein. Internal labels generally display less conformational flexibility than terminal labels, providing more precise positional information. Automated methods are used to rapidly generate assemblies of unique 3D models allowing the attachment sites of labeled domains to be accurately identified and thus provide an overall architectural map of the molecule. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2216.map.gz emd_2216.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2216-v30.xml emd-2216-v30.xml emd-2216.xml emd-2216.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  image2216.png image2216.png | 72 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2216 http://ftp.pdbj.org/pub/emdb/structures/EMD-2216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2216 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2216.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2216.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNaseIIIb-labeled Dicer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dicer labeled with Streptavidin at the RNase IIIb domain
| Entire | Name: Dicer labeled with Streptavidin at the RNase IIIb domain |
|---|---|
| Components |
|
-Supramolecule #1000: Dicer labeled with Streptavidin at the RNase IIIb domain
| Supramolecule | Name: Dicer labeled with Streptavidin at the RNase IIIb domain type: sample / ID: 1000 / Details: The sample was mostly monodisperse Oligomeric state: One human Dicer and one streptavidin molecule Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 280 KDa |
-Macromolecule #1: Human Dicer
| Macromolecule | Name: Human Dicer / type: protein_or_peptide / ID: 1 / Name.synonym: hDicer / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 220 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Macromolecule #2: Streptavidin
| Macromolecule | Name: Streptavidin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
| Molecular weight | Theoretical: 60 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 150mM KCl, 25mM HEPES |
|---|---|
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein floated on 2% w/v uranyl acetate |
| Grid | Details: holey C-flat grids covered with an additional layer of thin carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 62000 Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 50 |
| Date | Jan 10, 2011 |
| Image recording | Number real images: 880 / Average electron dose: 20 e/Å2 |
| Tilt angle min | 0 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each micrograph |
|---|---|
| Final two d classification | Number classes: 1 |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Software - Name: Spider / Number images used: 27607 |
| Details | Processing was done using APPION pipeline |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: Rigid body. manual docking based upon overall fit and correspondence to tagging data |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller