+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2198 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
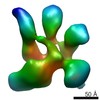
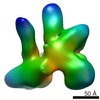
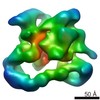
| Title | Architecture of human translation initiation factor 3 | |||||||||
 Map data Map data | reconstruction using Maximum-likelihood | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  translation initiation / translation initiation /  eukaryotic initiation factor 3 / eukaryotic initiation factor 3 /  ribosome / ribosome /  proteasome / PCI-MPN domains proteasome / PCI-MPN domains | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / cryo EM /  negative staining / Resolution: 20.0 Å negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Querol-Audi J / Sun C / Mortimer S / Arias-Palomo E / Vogan J / Doudna J / Nogales E / Cate J | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2013 Journal: Nucleic Acids Res / Year: 2013Title: Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Authors: Chaomin Sun / Jordi Querol-Audí / Stefanie A Mortimer / Ernesto Arias-Palomo / Jennifer A Doudna / Eva Nogales / Jamie H D Cate /  Abstract: The initiation of protein synthesis plays an essential regulatory role in human biology. At the center of the initiation pathway, the 13-subunit eukaryotic translation initiation factor 3 (eIF3) ...The initiation of protein synthesis plays an essential regulatory role in human biology. At the center of the initiation pathway, the 13-subunit eukaryotic translation initiation factor 3 (eIF3) controls access of other initiation factors and mRNA to the ribosome by unknown mechanisms. Using electron microscopy (EM), bioinformatics and biochemical experiments, we identify two highly conserved RNA-binding motifs in eIF3 that direct translation initiation from the hepatitis C virus internal ribosome entry site (HCV IRES) RNA. Mutations in the RNA-binding motif of subunit eIF3a weaken eIF3 binding to the HCV IRES and the 40S ribosomal subunit, thereby suppressing eIF2-dependent recognition of the start codon. Mutations in the eIF3c RNA-binding motif also reduce 40S ribosomal subunit binding to eIF3, and inhibit eIF5B-dependent steps downstream of start codon recognition. These results provide the first connection between the structure of the central translation initiation factor eIF3 and recognition of the HCV genomic RNA start codon, molecular interactions that likely extend to the human transcriptome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2198.map.gz emd_2198.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2198-v30.xml emd-2198-v30.xml emd-2198.xml emd-2198.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  2198_emd_2198.png 2198_emd_2198.png | 106.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2198 http://ftp.pdbj.org/pub/emdb/structures/EMD-2198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2198 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2198.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2198.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction using Maximum-likelihood | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.67 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PCI MPN core of the human eukaryotic translation initiation facto...
| Entire | Name: PCI MPN core of the human eukaryotic translation initiation factor eIF3 |
|---|---|
| Components |
|
-Supramolecule #1000: PCI MPN core of the human eukaryotic translation initiation facto...
| Supramolecule | Name: PCI MPN core of the human eukaryotic translation initiation factor eIF3 type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: Subcomplex containing 8 core subunits / Number unique components: 8 |
|---|---|
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: eIF3 a
| Macromolecule | Name: eIF3 a / type: protein_or_peptide / ID: 1 / Details: truncated version, contains residues 5-654 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #2: eIF3 c
| Macromolecule | Name: eIF3 c / type: protein_or_peptide / ID: 2 / Details: truncated version, contains residues 302-913 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #3: eIF3 e
| Macromolecule | Name: eIF3 e / type: protein_or_peptide / ID: 3 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #4: eIF3 f
| Macromolecule | Name: eIF3 f / type: protein_or_peptide / ID: 4 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #5: eIF3 h
| Macromolecule | Name: eIF3 h / type: protein_or_peptide / ID: 5 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #6: eIF3 k
| Macromolecule | Name: eIF3 k / type: protein_or_peptide / ID: 6 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #7: eIF3 l
| Macromolecule | Name: eIF3 l / type: protein_or_peptide / ID: 7 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #8: eIF3 m
| Macromolecule | Name: eIF3 m / type: protein_or_peptide / ID: 8 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20mM Hepes, 75mM KCl, 0.5mM EDTA, 1mM DTT, 2mM MgCl2, 3% trehalose |
| Staining | Type: NEGATIVE / Details: vitrified |
| Grid | Details: C-flat plasma cleaned with Solarus |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 100000 Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Date | Oct 9, 2011 |
| Image recording | Average electron dose: 20 e/Å2 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each image |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Software - Name: ML3D / Number images used: 32227 |
| Details | automatic particle selection with template correlator |
 Movie
Movie Controller
Controller